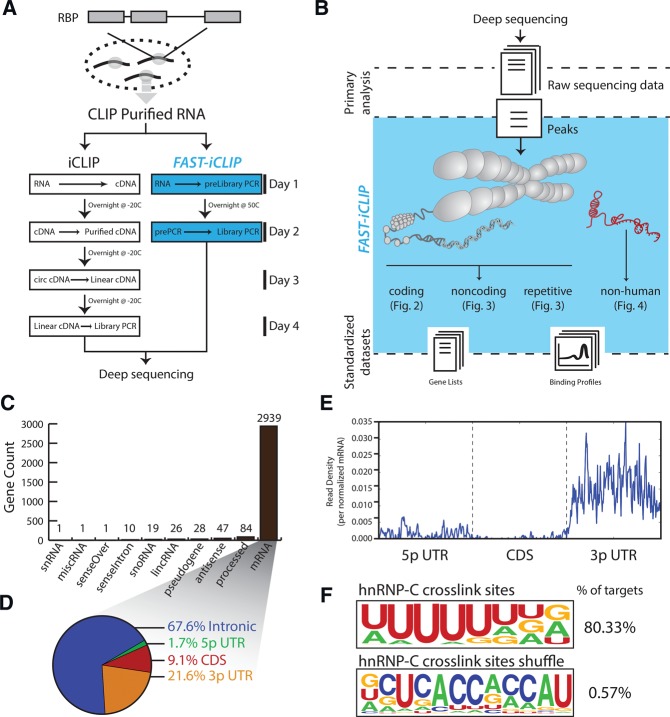
FIGURE 1.
FAST-iCLIP incorporates experimental improvements and standardized experimental interface to enable iCLIP analysis. (A) Biochemical improvements to the standard iCLIP procedure reduce experimental time by half. (B) A standard interface for analysis iCLIP data increases analysis efficiency and dissects many known sources of RNA transcripts including both the repetitive and nonrepetitive human genome as well as nonhuman genomes. (C) Histogram of the types and number of genes identified by FAST-iCLIP analysis of publically available hnRNP-C iCLIP data. (D) Percentage of all hnRNP-C iCLIP reads mapping to mRNA loci subdivided by functional domain. (E) Average histogram of hnRNP-C iCLIP reads along a normalized mRNA transcript. Each gene's functional regions (5′ UTR, CDS, and 3′ UTR) are binned into 200 units plotted along the same axis. Intronic reads are not visualized in this plot. (F) Logo visualization of the HOMER motif output from all hnRNP-C iCLIP reads with the fraction of iCLIP target regions containing that motif. hnRNP-C crosslink sites and region-shuffle control are shown.