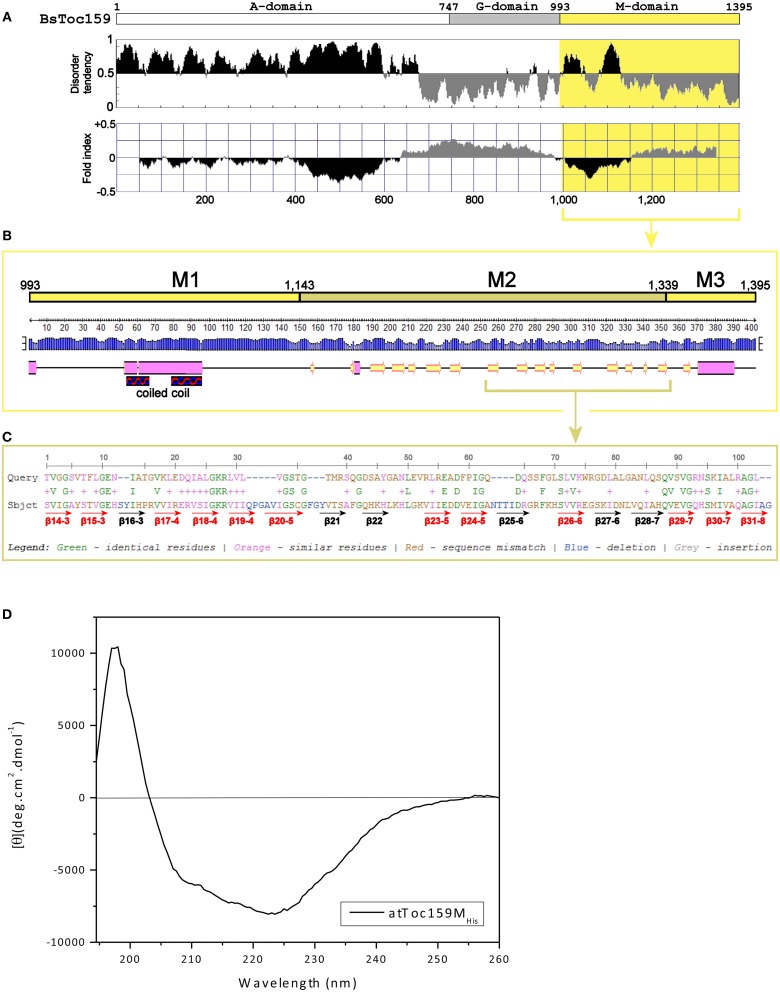
Figure 5.
Prediction of the M-domain structure. (A) Prediction of structurally disordered regions of BsToc159. IUPred (Dosztanyi et al., 2005; upper panel) and FoldIndex (Prilusky et al., 2005; lower panel) were used to predict the intrinsically disordered (shaded in black) and structured (shaded in gray) regions of the entire BsToc159 protein. The M-domain region is shaded in yellow. (B) Secondary structure prediction of the BsToc159 M-domain. Predictions were performed using the PSIPRED protein structure prediction server v3.0 (Jones, 1999). The height of the blue bar for each residue represents the confidence level. Cylinders, arrows and lines symbolize α-helices, β-strands and coils, respectively. Based on the structural prediction, the M-domain is subdivided into the M1, M2 and M3 subdomains: M1 represents a moderately disordered region with a putative α-helical region, which was predicted to fold into a coiled-coil structure by COILS (Lupas et al., 1991); M2 represents a β-strand-rich region; M3 represents the C-terminal 56-residue sorting signal. (C) Amino acid sequence alignment of M2 region with the lipid-binding domain of UDP-3-O-acyl-glucosamine N-acyltransferase (LpxD). BLAST search was performed using the AtToc159 M-domain as query sequence against 3D structures deposited in the RCSB Protein Data Bank (http://www.pdb.org/pdb/search/searchSequence.do). The β-strands which constitute the left-handed β-helix of LpxD (Buetow et al., 2007) are annotated by red (if homologous to M2) and black arrows (otherwise). (D) Far-UV CD spectrum of purified recombinant AtToc159MHis. The protein concentration was 9 μM. MRE is the mean residue ellipticity in degrees cm2 dmol−1. Temperature was 25°C.