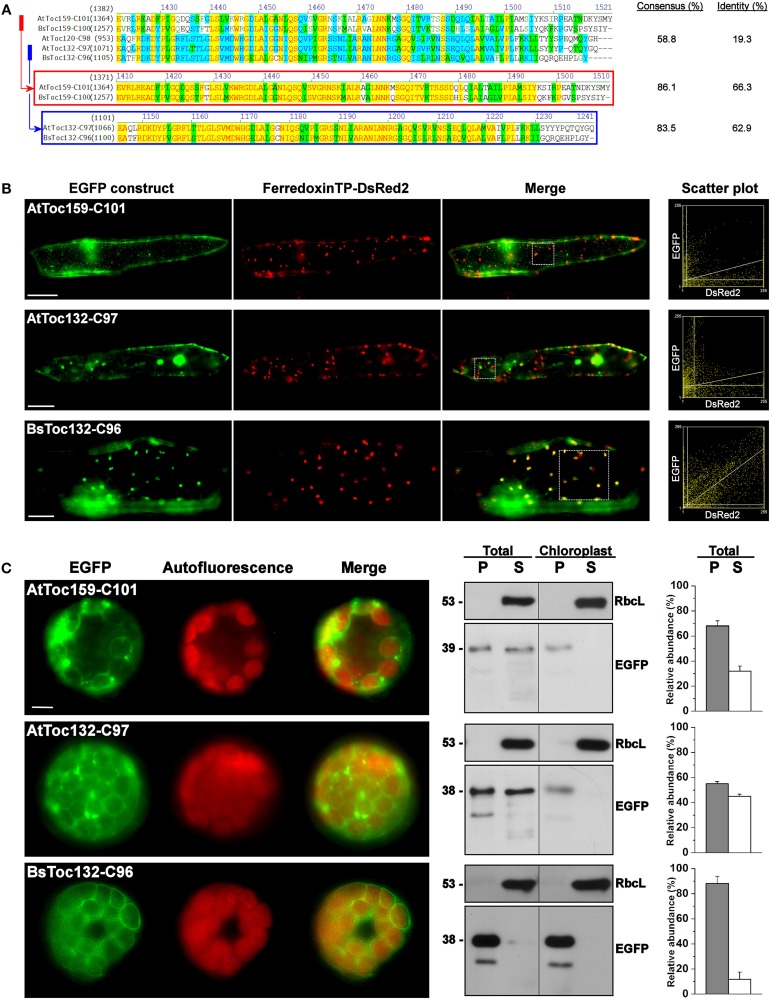
Figure 6.
Transient expression of EGFP fusion proteins with the C-terminal regions of other Toc159 homologs. (A) Amino acid sequence alignment of the Toc159 homologs from B. sinuspersici and A. thaliana. Sequence homologies among the Toc159 and Toc132 isoforms are shown in red and blue boxes, respectively. Alignment was performed using the AlignX module of Vector NTI Advance™ 10.3.0 (Invitrogen) and is displayed using the default color scheme: a red foreground on a yellow background denotes a 100% conserved residue; a dark green foreground on a white background denotes a residue with weak similarity to the consensus residue at a given position; a black foreground on a light green background denotes a consensus residue in a block of similar residues at a given position; A blue foreground on a cyan background denotes a conserved residue with 50% or higher identity at a given position; A black foreground on a white background denotes a non-similar residue. (B) Colocalization analysis of EGFP fusion proteins in onion epidermal cells. EGFP was fused to the C-terminal regions of other Toc159 homologs equivalent to the EGFP-BsToc159-C100 construct based on the protein alignment as shown in (A). Details are the same as in Figure 2. (C) Transient expression of EGFP fusion proteins in isolated A. thaliana protoplasts. Details are the same as in Figure 3.