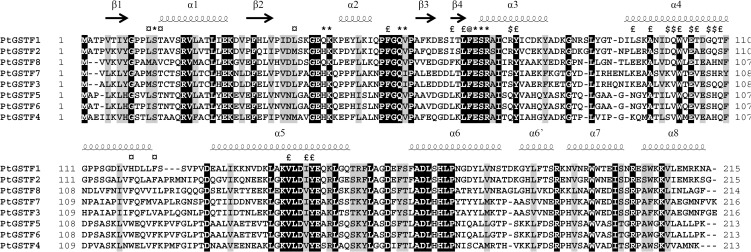
Figure 1.
Structure based sequence alignment of Populus trichocarpa GSTFs. The structural alignment was calculated with PROMALS3D using the structure of PttGSTF1 and rendered using ESpript 3.0. Accession numbers of GSTFs from the version 3.0 of P. trichocarpa genome are the following: PtGSTF1: Potri.002G015100, PtGSTF2: Potri.002G015200, PtGSTF3: Potri.014G132200, PtGSTF4: Potri.T035400, PtGSTF5: Potri.T035300, PtGSTF6: Potri.T035100, PtGSTF7: Potri.T035000, PtGSTF8: Potri.017G138800. From the structure of PttGSTF1, residues contributing to the dimerization and those involved in the G- or H-sites are highlighted as follows: *, glutathione-interacting residues; ¤, MES-interacting residues; $, dimer interface via hydrogen bond; £, dimer interface via van der Waals interactions; @, dimer interface via hydrogen bond and van der Waals interactions.