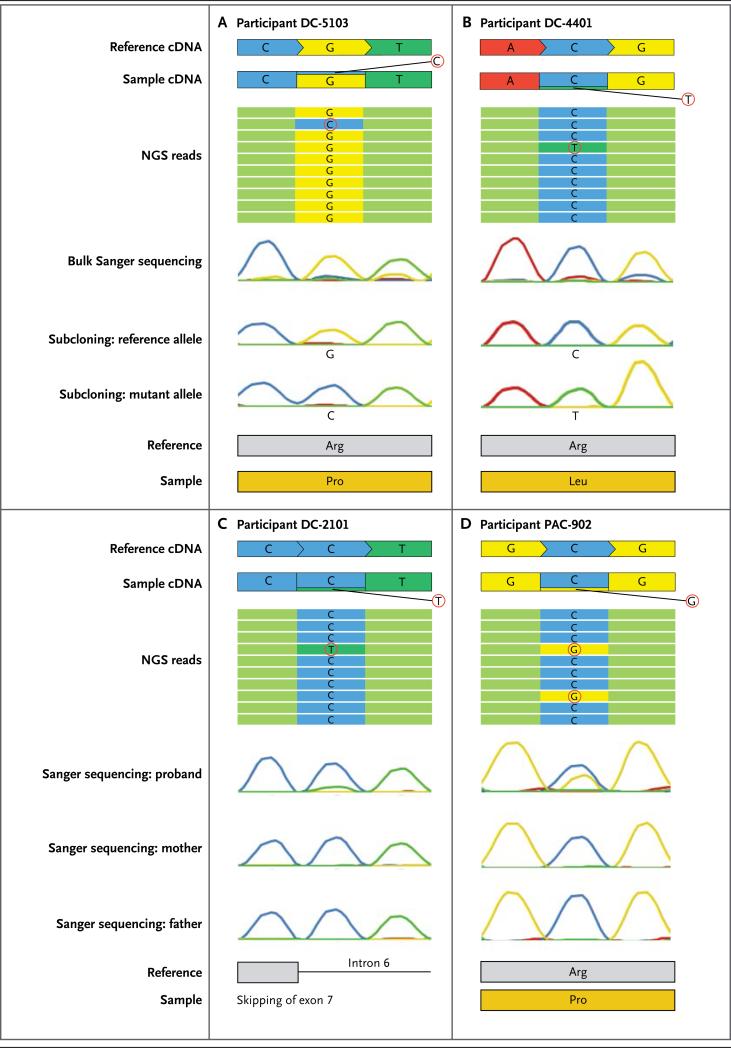
Figure 2. Detection of Variants by Means of NGS and Sanger Sequencing.
The figure shows NGS reads aligned with Sanger se- quencing chromatograms from bulk DNA amplifica- tion and after subcloning for Participant DC-5103 (Panel A) and Participant DC-4401 (Panel B); the vari- ants (circled nucleotides) were detected in a fraction of the reads but were missed when targeted Sanger sequencing was used. Subcloning detected the vari- ants in a proportion of clones, and the proportion of NGS reads was strongly correlated with the propor- tion of clones containing a mosaic read. NGS reads aligned with Sanger sequencing chromatograms from bulk DNA amplification of samples from Participant DC-2101 (Panel C) and Participant PAC-902 (Panel D) and their respective parents show that the mosaic variant (circled nucleotide) was detected as a smaller peak, as compared with the reference allele, on Sanger sequencing and arose de novo (i.e., it was absent from the parents). For the splicing mutation in Participant DC-2101, the prediction software did not enable a choice from among the potential splice-acceptor sites; therefore, no amino acid is depicted.