Figure 1.
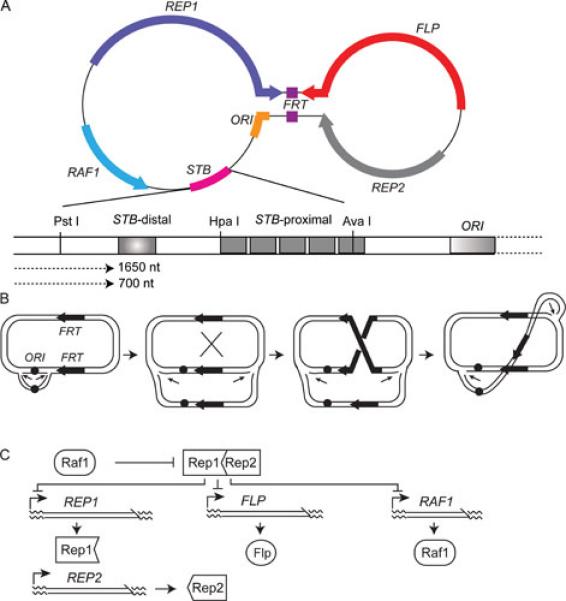
Genetic organization of the yeast plasmid and its copy number regulation. The double stranded DNA genome of the yeast 2 micron plasmid is generally represented as a dumbbell shaped molecule to highlight the 599 bp inverted repeat (the handle of the dumbbell) that separates two unique regions. The four coding regions harbored by the plasmid are REP1, REP2, FLP and RAF1. The directions in which these loci are transcribed are indicated by the arrowheads. The plasmid replication origin is indicated as ORI. Flp is a site-specific recombinase, whose target sites (FRTs) are embedded within the inverted repeat region. The plasmid partitioning locus STB can be divided into origin-proximal (STB-proximal) and origin-distal (STB-distal) segments. There are five repetitions of a 60 bp consensus sequence in STB-proximal. STB-distal, which harbors the termination signal for two origin directed plasmid transcripts (1660 nt and 700 nt long) as well as silencing element (shaded box), maintains STB-proximal as a transcription-free zone. The Rep1-Rep2-STB system ensures equal plasmid segregation. The (Flp-FRT)-Raf1 system is responsible for the maintenance of steady state plasmid copy number. B. The mechanism proposed for copy number correction of the plasmid by amplification (12) invokes a Flp mediated recombination event that changes the direction of one of the replication forks (indicated by the thin short arrows) with respect to the other during bidirectional replication of the plasmid. The ensuing dual uni-directional mode of replication amplifies the plasmid as a concatemer of tandem plasmid units. There is a marked asymmetry in the location of the FRT sites (thick arrows) with respect to the replication origin (ORI). The consequent difference in their replication status, one duplicated and the other not, is responsible for the relative inversion of the replication forks as a result of recombination between them C. Efficient amplification without the danger of unregulated increase in plasmid copy number is prevented by a transcriptional regulatory network. The putative [Rep1-Rep2] repressor negatively regulates FLP, RAF1 and REP1 expression. Raf1 is thought to antagonize the action of the [Rep1-Rep2] repressor.
