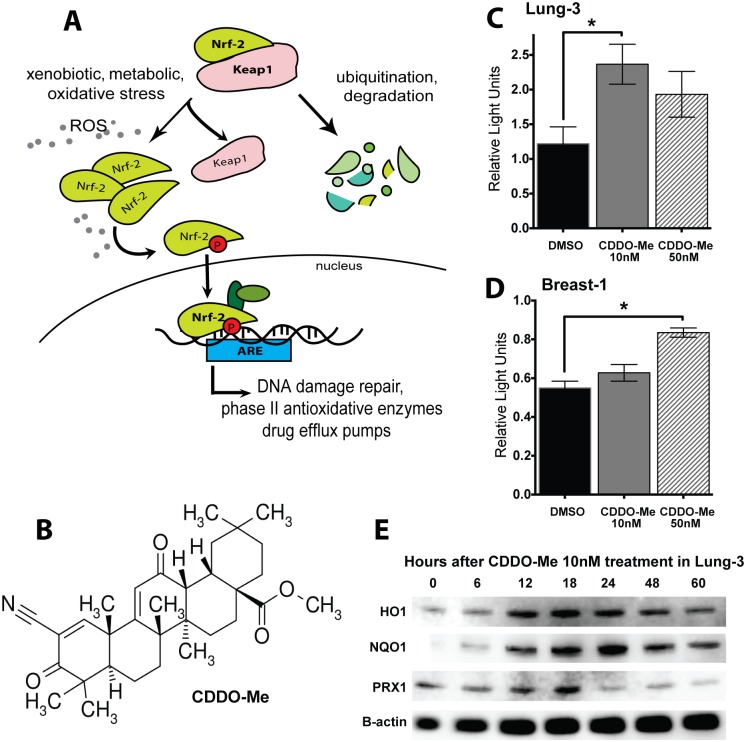
Figure 1. CDDO-Me activates the Nrf2 antioxidant pathway in epithelial cells.
(A) Nrf2 Pathway: Nrf2 is a transcription factor normally bound by its cytoplasmic repressor Keap1, which acts as a molecular oxidative sensor and marks Nrf2 for degradation. When there is an abundance of reactive species in the cells, Nrf2 accumulates in the cytoplasm, eventually undergoing various phosphorylation events to translocate to the nucleus and bind to Antioxidant Response Elements (AREs) in the genome, resulting in the transcription of multiple antioxidative and cyto-protective genes. CDDO-Me acts by facilitating the dissociation between Keap1 and Nrf2, leading to Nrf2 activation. (B) Chemical structure of CDDO-Me: Oleana-1,9(11)-dien-28-oicacid, 2-cyano-3,12-dioxo-, methyl ester (RTA-402; bardoxolone-methyl). (C, D) CDDO-Me increases expression of ARE-driven luciferase 18 hours after drug treatment in HBEC 3KT and HME1, respectively. Firefly ARE-luciferase normalized to renilla control (RLU). Mean ± SEM of 6 replicates, *p<0.05 using paired t-test (between DMSO and drug). (E) CDDO-Me 10 nM activates heme oxygenase-1 (HO1, band observed at ∼32 kDa), NADPH dehydrogenase quinone (NQO1, band observed at ∼30 kDa), and peroxiredoxin (PRX1, band observed at ∼20 kDa), all downstream targets of Nrf2/ARE and peaking at approximately18 hours after treatment in HBEC 3KT.