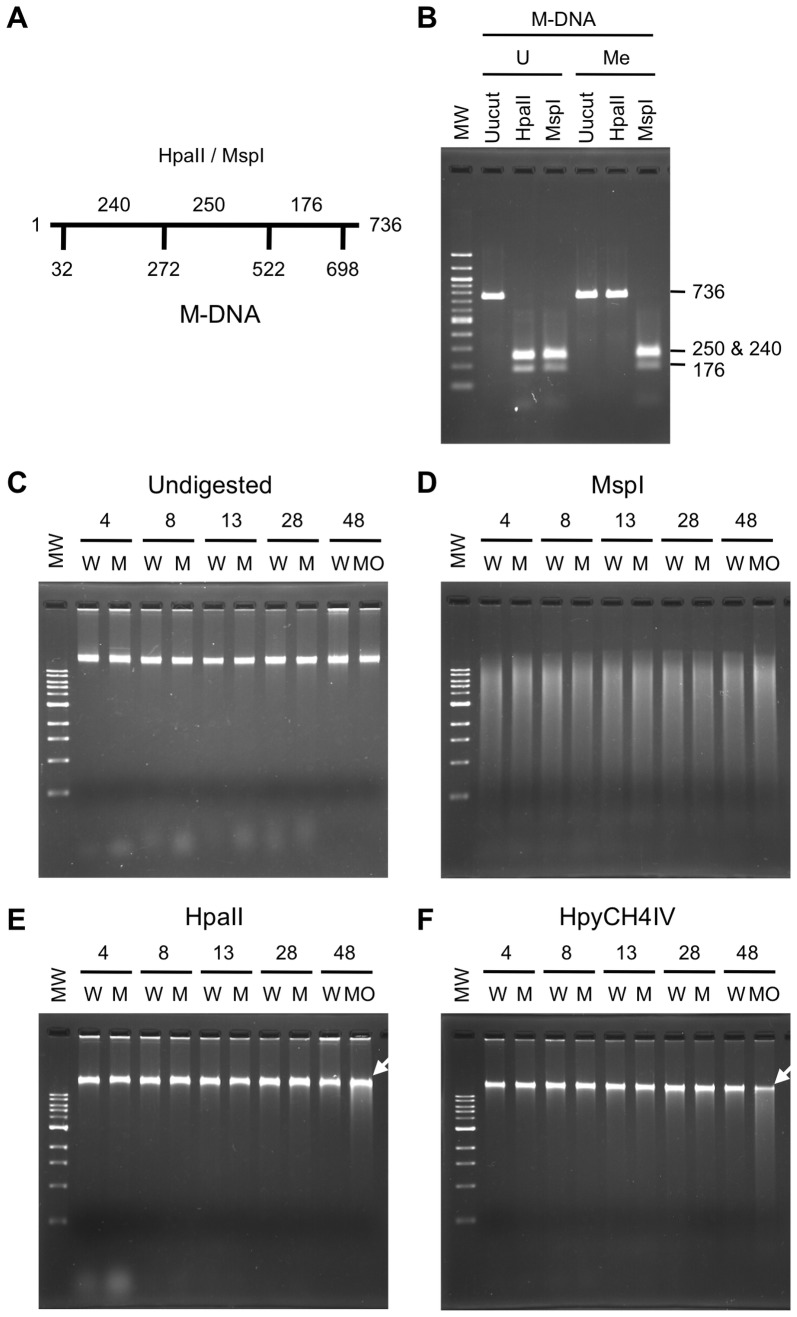
Figure 1. Absence of genome demethylation following the injection of a methylated DNA fragment into zebrafish eggs.
(A) A schematic structure of the 736-bp DNA template used for in vitro methylation by HpaII methylase to generate M-DNA. Numbers below the horizontal line indicate the positions of HpaII/MspI sites subject to methylation. Numbers above the line show the lengths of the fragments generated when completely digested with HpaII or MspI. (B) Verification of in vitro methylation of the 736-bp DNA template. The unmethylated DNA fragment (U) was susceptible to HpaII, a methylation-sensitive restriction enzyme, as well as MspI, the methylation-insensitive isoschizomer of HpaII. Methylation of the fragment (Me) conferred resistance to DNA to the digestion by HpaII. (C) Undigested genomic DNA of control (W), M-DNA-injected (M), and dnmt1 MO-injected (MO) embryos at the indicated time points (hours post fertilization; hpf) were run on an agarose gel. (D, E, F) The same genomic DNAs as those used and shown in (C) were digested with MspI (D), HpaII (E), or HpyCH4IV (F), and run on an agarose gel. The molecular weight markers of DNA loaded on the first lanes of gels, shown as MW, were 100 bp (B) or 1 kb ladders (C, D, E, and F). Note that smearing down of high-molecular DNA, thereby reducing the methylation level, was discernible only in the genomic DNA from dnmt1 MO-injected embryos (white arrows in E and F).