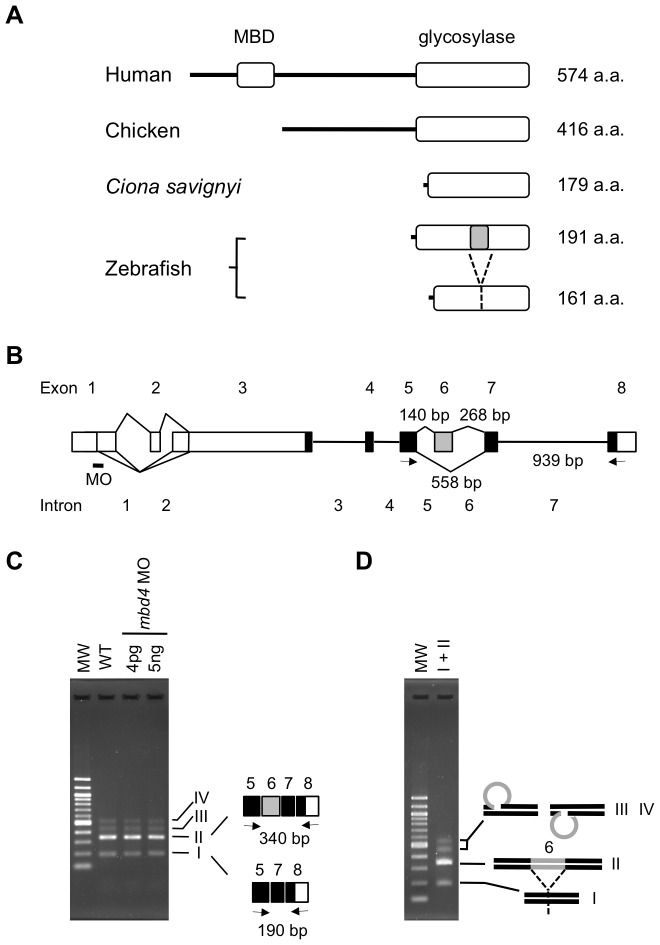
Figure 4. Zebrafish MBD4 lacks a MBD and mbd4 MO did not interfere splicing of transcripts.
(A) Schematic structures of MBD4 proteins in the chordata are indicated. The predominant form of MBD4 had MBD and a glycosylase domain near the N- and C- termini, respectively, similar to human MBD4. As rare cases in chordate, such as Chicken and Ciona, two zebrafish MBD4 proteins generated via alternative splicing lacked MBD. The shorter isoform lacked thirty amino acids, which corresponded to exon 6 (marked in grey). (B) The schematic structure of the zebrafish mbd4 gene. Boxes show exons, and black and white boxes show translated and untranslated regions, respectively. Alternative splice events are indicated as diagonal lines. Five splicing variants of the 5′UTR and a shorter isoform that skipped exon 6 (grey box) were obtained by RT-PCR. The approximate positions of MO and primer pairs designed by Rai et al. [3] to detect aberrant splicing by MO are indicated by a small bar and arrows, respectively. The lengths of introns that may have been included in the RT-PCR product are shown. (C) With the primer set shown in (B), a region of mbd4 cDNA was amplified from wild-type (WT) embryos in which the indicated amounts of mbd4 MO were injected. The same banding pattern was observed irrespective of the MO injection. In addition to the expected band (II), three faint, but distinct bands (I, III, and IV) appeared both in wild-type and MO-injected embryos. The bottom band (I) was derived from an alternative splicing variant that skipped exon 6. (D) PCR amplification of band I and II in a single tube reproduced the same banding pattern on agarose gel as that in (C). The possible structures of bands III and IV are shown on the right of the gel.