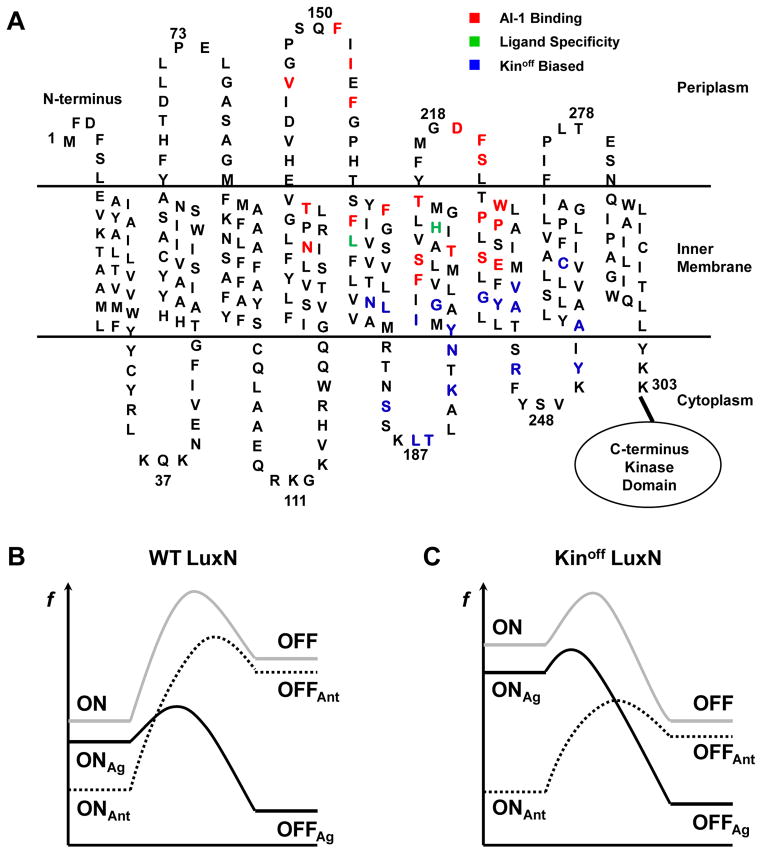
Figure 7. Sites of LuxN mutants with altered ligand specificity and a proposed model for LuxN agonism and antagonism.
(A) Predicted topology of the LuxN transmembrane domain. Residues required for AI-1 ((R)-3OH-C4 HSL) detection are shown in red. Leu166 and His210 that determine ligand specificity are shown in green. Kinoff biased mutation sites are shown in blue.
(B) An energy profile illustrating theoretical transitions of the WT LuxN receptor between the Kinon and the Kinoff states upon ligand binding. Notations are: f (free energy), ON (unliganded Kinon state), OFF (unliganded Kinoff state), ONAg (agonist-bound Kinon state), OFFAg (agonist-bound Kinoff state), ONAnt (antagonist-bound Kinon state), OFFAnt (antagonist-bound Kinoff state). Grey solid, unliganded; black solid, agonists bound; dotted black, antagonists bound.
(C) An energy profile illustrating the analogous transitions for the Kinoff biased LuxN receptor. Notations are as in B.