Figure 2.
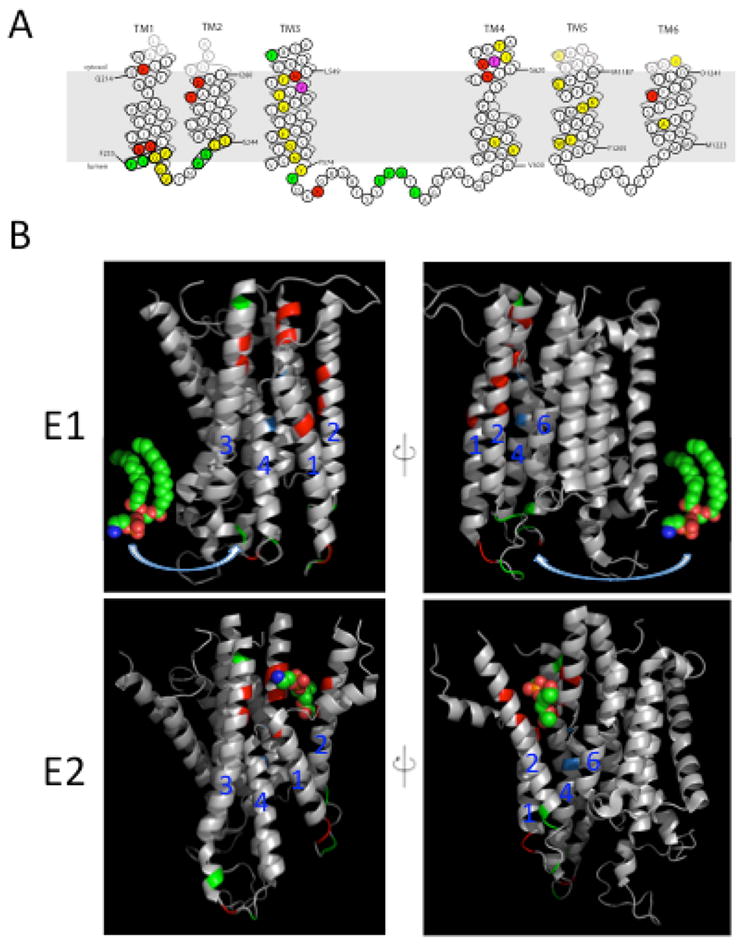
Amino acid residues in the membrane domain of Dnf1 that determine substrate specificity. (A) Topology of the first 6 transmembrane (TM) segments of Dnf1 highlighting residues important for substrate specificity and transport. Dnf1 normally recognizes and transports lyso-PC and lyso-PE, but specific point mutations changing the residues highlighted in red allow recognition of PS. Mutations in residues highlighted in green perturb recognition of PC without altering translocation of PE. Changes in only two residues, highlighted in purple, alter recognition of both PC and PS. Mutation of residues in yellow reduced activity without altering substrate specificity. (B) Two potential translocation pathways for flipping phospholipid substrate. The phospholipid headgroup may engage the entry gate residues in the E1 conformation, then slide along a grooved formed by TM segments 1, 3 and 4 (left) or TM 2, 4 and 6 (right) and dock in the exit gate in the E2 conformation. The crystal structures of the Ca2+-ATPase membrane domain in the E1 (PBD ISU4) and E2 (PDB 3W5C) conformational states are shown with the positions homologous to substrate-defining residues of the P4-ATPases highlighted. Red and green positions represent the Dnf1 residues shown in (A) involved in PS and PC recognition, respectively. Positions highlighted in blue represent the P+1 Ile and N359 in ATP8A2. The PE molecule in the E1 images derived from PDB 3B74 while the PE headgroup in the E2 images co-crystallized with the Ca2+-ATPase.
