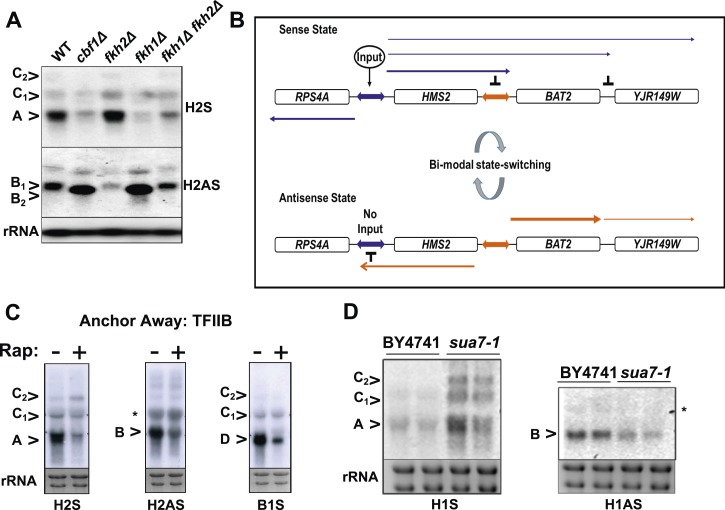
Figure 6. General and specific transcription factors control state switching at HMS2.
(A, C, D) Northern blots showing sense or antisense-dominant state at HMS2 in strains lacking general or specific transcription factors. (A) Strains were cultured in YPD depleted for tryptophan. (B) Model for state-switching between a sense-dominant state and an antisense-dominant state by transcription factors (Input) at the divergent promoters between RPS4A and HMS2 (blue double arrow) and HMS2 and BAT2 (orange double arrow). During growth in glucose (high input), cells cycle between the sense-dominant state and the antisense-dominant state with the majority of cells existing in the sense-dominant state. During growth in galactose (low input), cells cycle between the sense-dominant state and the antisense-dominant state with the majority of cells existing in the antisense-dominant state. (C) Anchor Away of TFIIB (Sua7) is achieved after incubation with rapamycin (Rap) (+) in DMSO for 1 h or DMSO alone (−). (D) Biological replicates for the WT strain BY4741 and the isogenic strain expressing the sua7-1 allele are shown.