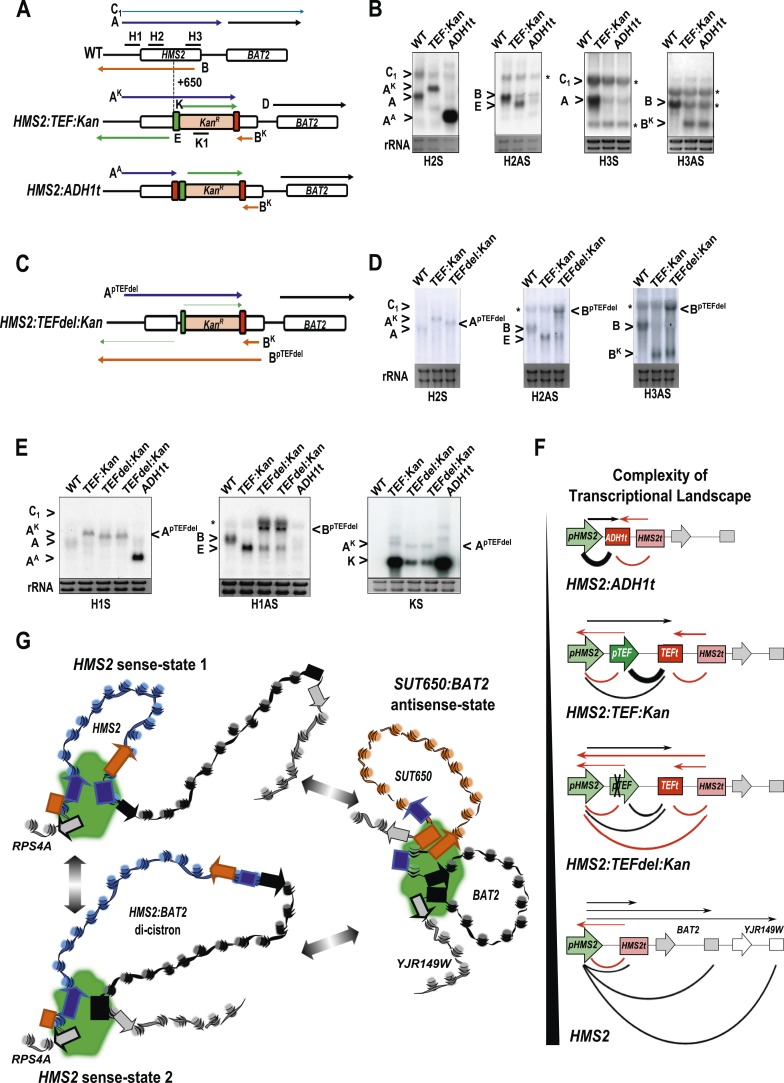
Figure 7. The plasticity of transcription units.
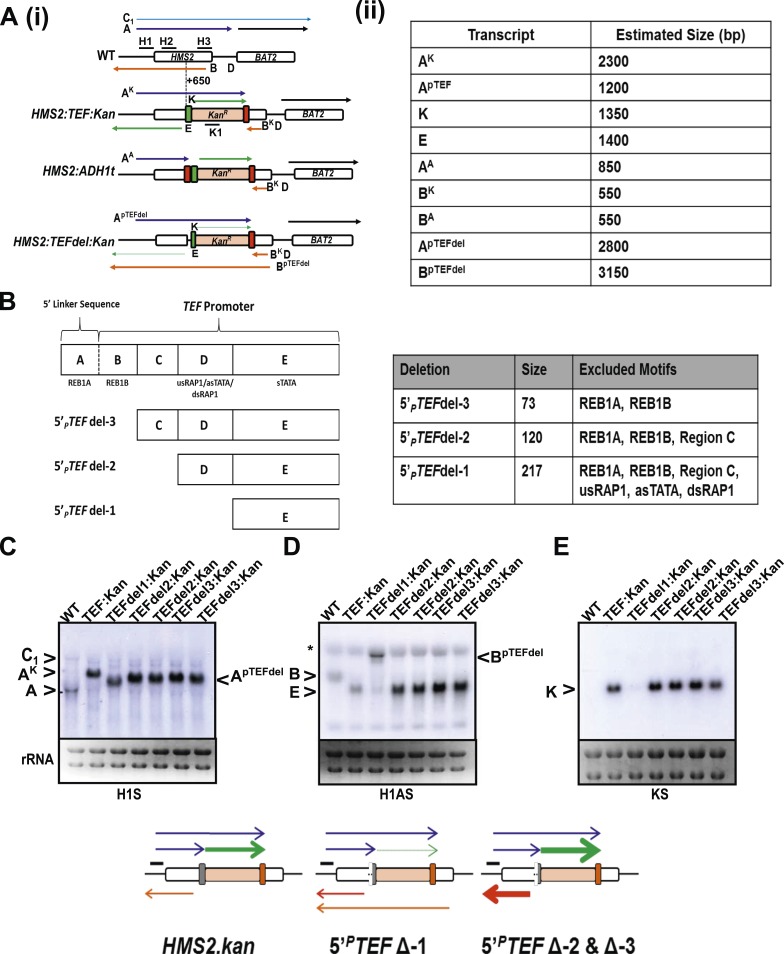
(A, C) Schematic of constructs showing transcripts and position of probes. Transcripts resulting from the pTEF:kanMX:TEFt insertion, the same construct with the ADH1 terminator (pink box), or the TEF:kanMX:TEF insertion with a deleted TEF promoter (p) are identified with a superscript K, A, or pTEFdel, respectively (see Figure 7—figure supplement 1 for more details). (B, D, E) Northern blots in the strains indicated showing the conditional nature of the TEF terminator (t) when the pTEF is compromised. (F) Modelling transcription in an interleaved genome demonstrating an increase in complexity of the transcriptional landscape as the strength of terminators and promoters decreases. Constructs representing each state are indicated; grey and white boxes are downstream TUs. Solid directional boxes are promoters; squares are terminators. Deeper colour indicates increased strength. Arrows on top of diagrams represent the transcripts initiating or terminating at pHMS2 and HMS2t (black sense; red antisense). Curved lines under diagrams show the extent of each individual transcription unit. Transcription units are envisaged as dynamic with any one region only involved in one TU at any time in an individual cell (see Figure 7G). Weakening of pTEF in the HMS2:TEF:Kan construct results in a level of transcription complexity (number of different TUs) similar to the native HMS2 locus. (G) Schematic showing the dynamic alternative transcription units at the HMS2:SUT650:BAT2 locus. Directional boxes are promoters, squares are terminators coloured coded according to the nature of the TU to which they belong (blue, HMS2; black, BAT2; Orange, SUT650; Grey, RPS4A and YJR149W). HMS2 forms alternative TUs with its own terminator (sense-state 1) or with the BAT2 terminator (sense-state 2—a di-cistronic transcription unit) excluding the SUT650, BAT2, and YJR149W promoters from a region where transcription can occur (green shading). RPS4A, an OX gene expressed divergently from HMS2 is active. The sense state predominates in the OX phase of the YMC and during growth on GLU. Any promoter or terminator excluded from the transcription machinery is neutral to the transcription process, explaining how both promoters and terminators are subject to extensive read-through transcription. The sense-state toggles to the alternative antisense-state, predominant in GAL and during the RC phase of the YMC. Here, the formation of the SUT650 transcription unit excludes the HMS2 and RPS4A promoters from the transcription machinery. This relieves interference of BAT2 and YJR149W by HMS2 sense transcription. One transcription unit is envisaged to exist in one cell at any one time. The antisense state is the default, requiring transcription factor-dependent activation at HMS2 to switch to the sense state.