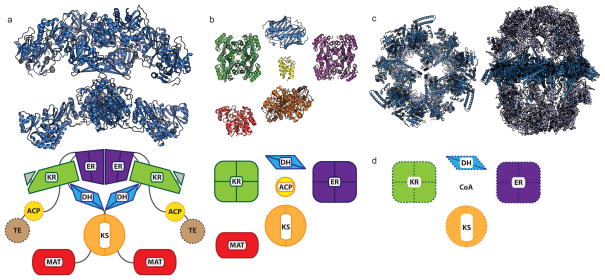
Figure 1.
Comparison of FAS domain organizations. a) Mammalian type I synthase (PDB: 2VZ8) structure with a diagram below of the domain organization. Domains that were not observed in the crystal structure are denoted by hashed borders. b) E. coli type II discrete structures with a comparative diagram below denoting multimeric states of the proteins. (PDB: KR, 1Q7B; DH, 1MKB; ER, 1DFI; ACP, 2FAD; KS, 2VB9; MAT, 2G2Z) c) Two fungal type I synthases demonstrating increased scaffolding elements and organization compared to the mammalian type I synthase in (a). At left, top-view cross-section of FAS from yeast Saccharomyces cerevisiae (PDB: 2UV8). At right, thermophilic fungus Thermomyces lanuginosus (PDB: 2UVB, 2UVC). d) Domain organization for the ACP-independent type II-like FAS discovered in Archaea, from which no structures are currently available.