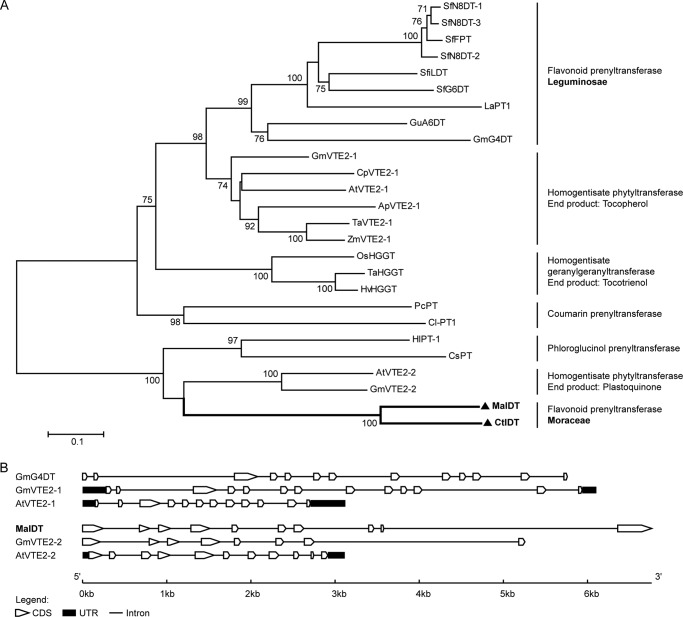
FIGURE 5.
Phylogenetic and genome structure analyses of MaIDT and CtIDT. A, the phylogenetic relationship between the MaIDT and CtIDT proteins and the related plant prenyltransferases. The protein sequences were aligned using ClustalW. The neighbor-joining phylogenetic tree was drawn using MEGA6. The bootstrap value was 1000, and the branch lengths represent the relative genetic distances. The abbreviations of the protein sequences and their accession numbers are as follows: MaIDT (M. alba; KM262659); CtIDT (C. tricuspidata; KM262660); SfFPT (S. flavescens; KC513505); SfN8DT-1 (S. flavescens; AB325579); SfN8DT-2 (S. flavescens; AB370330); SfN8DT-3 (S. flavescens; AB604222); SfG6DT (S. flavescens; BAK52291); SfiLDT (S. flavescens; AB604223); LaPT1 (L. albus; JN228254); GmG4DT (G. max; AB434690); GuA6DT (G. uralensis; KJ123716); ZmVTE2–1 (Zea mays; DQ231055); TaVTE2–1 (Triticum aestivum; DQ231056); ApVTE2–1 (Allium porrum; DQ231057); CpVTE2–1 (Cuphea pulcherrima; DQ231058); AtVTE2–1 (Arabidopsis thaliana; AY089963); GmVTE2–1 (G. max; DQ231059); PcPT (Petroselinum crispum; AB825956); Cl-PT1 (Citrus limon; AB813876) OsHGGT (Oryza sativa; AY222862); HvHGGT (Hordeum vulgare; AY222860); TaHGGT (T. aestivum; DQ231056); AtVTE2–2 (A. thaliana; DQ231060); GmVTE2–2 (G. max; DQ231061); HlPT-1 (Humulus lupulus; AB543053); CsPT (Cannabis sativa; see Ref. 51); OsPPT1 (O. sativa; AB263291); AtPPT1 (A. thaliana; AY089963); LePGT1 (Lithospermum erythrorhizon; AB055078); LePGT2 (L. erythrorhizon; AB055079). B, comparisons of the exon-intron structures of MaIDT and the related prenyltransferases. The gene sequence of MaIDT can be acquired from MorusDB with the gene ID Morus014065; the GenBankTM accession numbers of the other gene sequences used have been provided above. CDS, coding sequence; UTR, untranslated regions.