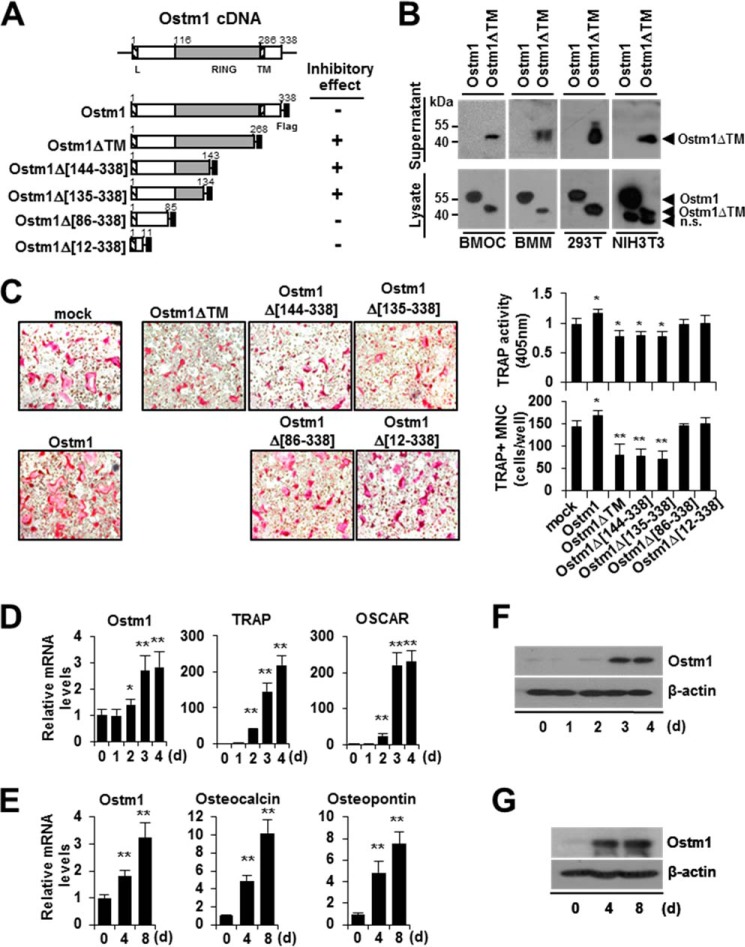
FIGURE 1.
Negative effects of the truncated OSTM1 mutant on osteoclastogenesis. A, schematic illustration of the murine Ostm1 cDNA structure and the OSTM1 expression plasmids. The putative RING domain (gray box) and leader/transmembrane sequences (hatched box) are indicated with amino acid residues. Flag, FLAG epitope (black box). The inhibitory effects of OSTM1 deletion mutants in osteoclastogenesis are summarized. B, extracellular secretion of truncated OSTM1 in BMOCs, BMMs, 293T, and NIH3T3 cells. Extracellular secretion of FLAG-tagged OSTM1ΔTM from culture supernatants of cells was detected through immunoblotting with antibodies against the FLAG epitope. n.s., nonspecific band. C, effect of truncated OSTM1 overexpression on OC differentiation. BMMs transduced with retroviral supernatants expressing OSTM1 or OSTM1 truncated mutants were cultured with RANKL and M-CSF. OCs were analyzed and photographed (left; original magnification, ×100) after TRAP staining. The TRAP solution assay was performed (top right panel). The number of TRAP+ MNCs (≥3 nuclei) was counted (bottom right panel). D, analysis of Ostm1 gene expression in BMOCs through real-time PCR. BM cells were differentiated into BMOCs using both M-CSF and RANKL stimulation. β-Actin expression served as an internal control. TRAP, tartrate-resistant acid phosphatase; OSCAR, OC-associated receptor. E, analysis of Ostm1 gene expression in pOBs through real-time PCR. Murine calvaria-derived stromal cells were differentiated into OBs for 8 days. F, immunoblot analysis of Ostm1 gene expression in BMOCs. BMOCs were prepared and analyzed by immunoblotting with specific antibodies. β-Actin was used as the loading control. G, immunoblot analysis of Ostm1 gene expression in pOBs. *, p < 0.05; **, p < 0.01. Error bars, S.D.