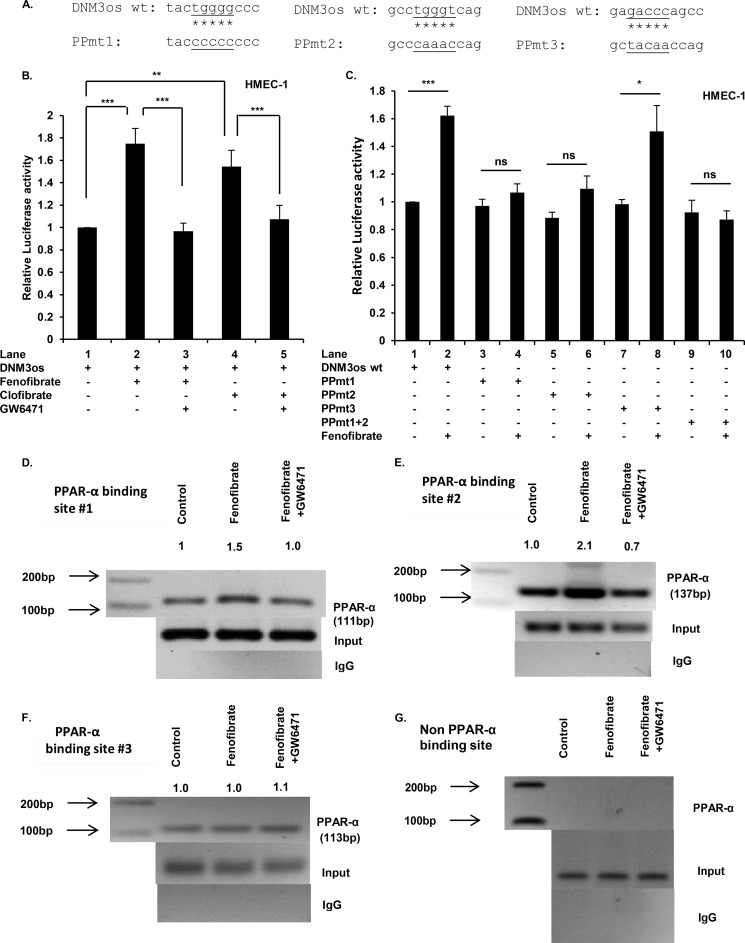
FIGURE 5.
PPARα cis-binding elements in promoter of DNM3os regulate expression of DNM3os as determined by reporter assay and ChIP. A, schematic of mutations in core sequences corresponding to PPARα sites (1, 2, and 3; see Fig. 4A) in DNM3os promoter reporter constructs. B and C, HMEC cells were transfected with either DNM3os-luc (WT) plasmid or the mutant plasmid with singly mutated PPARα binding sites (1, 2, or 3) or double mutation of PPARα sites 1 and 2 in DNM3os-luc constructs (PPmt). Transfected cells were treated as indicated with PPARα agonists (fenofibrate or clofibrate) or antagonist (GW6471) for 24 h. The cell lysates were assayed for luciferase activity and normalized to Renilla activity. The data are means ± S.E. for n = 3. D–F, HMEC cells were treated with either fenofibrate or fenofibrate +GW6471 for 2 h. The soluble chromatin was isolated and immunoprecipitated with either antibody to PPARα or control rabbit IgG. The PCR primers flank PPARα binding sites (1, 2, and 3), as indicated in schematic shown in Fig. 4A, and are listed in Table 1. Immunoprecipitated chromatin was processed for PCR analysis, and the expected product sizes are indicated. The middle panel shows amplification of the input DNA before immunoprecipitation. The data are representative of two independent experiments ran in duplicate, and the indicated values are densitometry values normalized to input DNA. G, immunoprecipitated chromatin was processed for PCR analysis utilizing primers corresponding to non-PPARα binding site in the DNM3os promoter. ns, not significant.