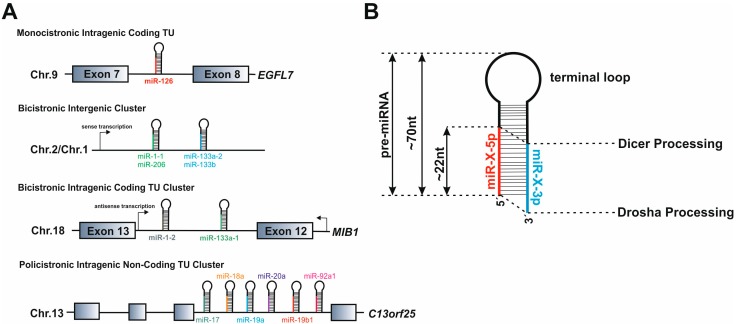
Figure 3.
miRNA genomic organization and pre-miRNA hairpin loop features. (A) Mammalian miRNA genes are encoded in defined transcription units (TUs) that based on their genomic localization can be classified into three major groups: (i) intronic miRNA genes in protein-coding genes; (ii) intergenic miRNA genes; (iii) intronic miRNA genes in non-coding TUs. miRNA genes can be clustered in a single polycistronic transcript that can be processed in order to generate two or more mature miRNAs. Some miRNA genes are located within TUs with the same transcription orientation as the host gene, whereas others can be transcribed in the anti-sense orientation. (B) miRNA precursors fold into a hairpin loop structure that is sequentially processed. Drosha cleaves the pri-miRNA at the base of the stem-loop, generating a pre-miRNA (~70 nt long) precursor that is further processed by Dicer in order to produce a miRNA double-stranded duplex (~22 nt). Both Drosha and Dicer processing generates a characteristic 2 nt 3'overhang. Depending on the relative localization in the hairpin-loop, the mature miRNA can be termed -5p or -3p.