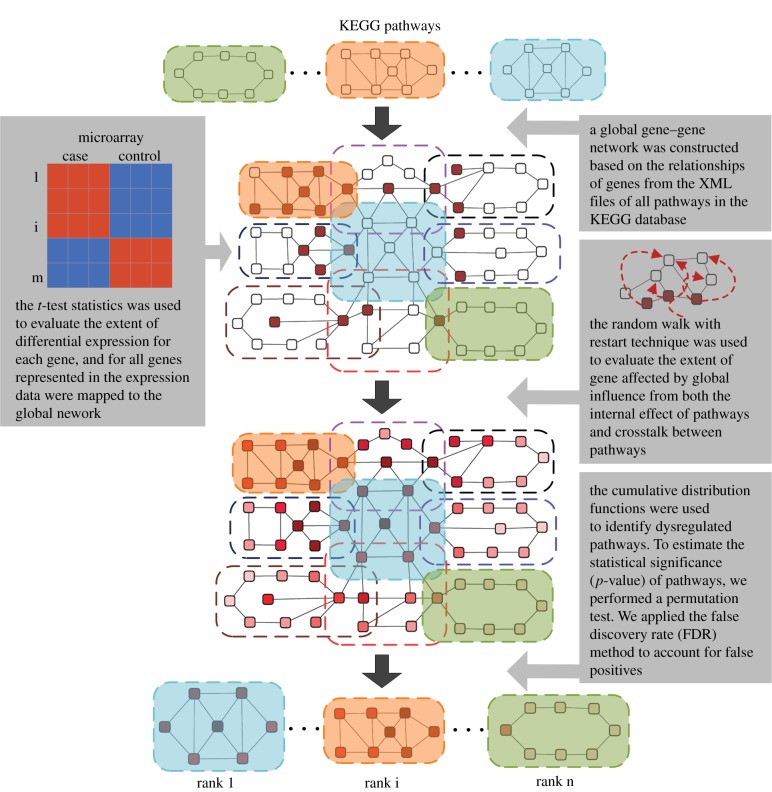
Figure 1.
Flow diagram of the methodology. A global gene–gene network was initially constructed on the basis of the relationships among genes extracted from all pathways. The extent of differential expression for each gene was evaluated by t-test, and all genes represented in the expression data were mapped to the global network. The RWR algorithm was then used to evaluate the number of genes affected by global influence from both internal effects of the pathways and crosstalk between pathways. Finally, cumulative distribution functions were used to identify dysregulated pathways. (Online version in colour.)