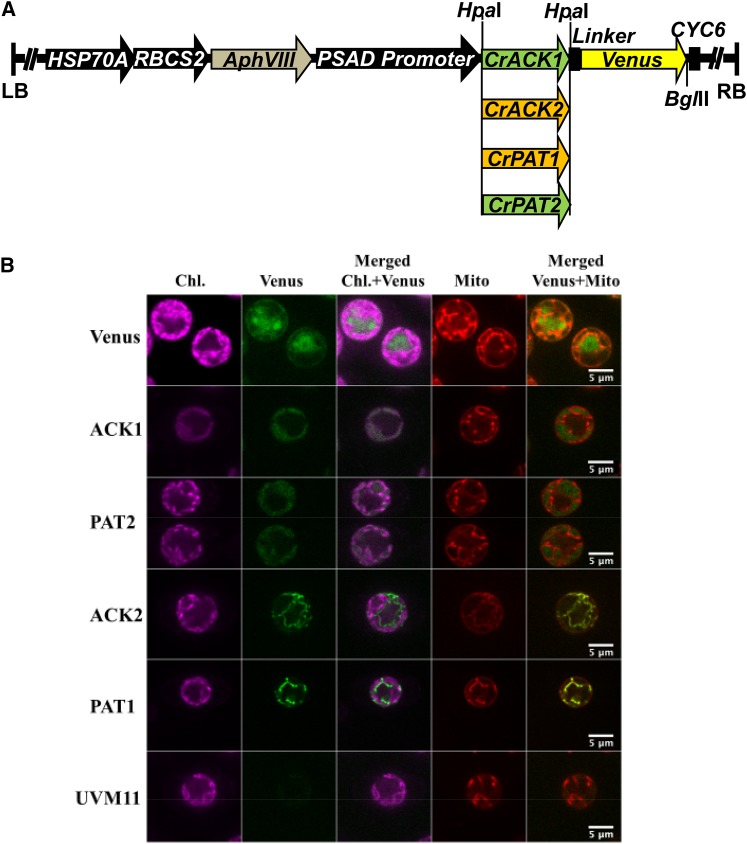
Figure 3.
Localization of ACK and PAT Isozymes.
(A) Constructs for expression of ACK or PAT fused to Venus. Black arrows represent promoters; AphVIII confers paromomycin resistance, and CYC6 is a terminator. LB and RB, left and right border, respectively. Green represents genes encoding predicted chloroplast-localized proteins, and brown denotes those predicted to be mitochondrial.
(B) Localizations of ACK and PAT Venus fusion proteins in Chlamydomonas. For the indicated proteins, chlorophyll autofluorescence (Chl.), Venus fluorescence, merged chlorophyll and Venus signals, fluorescence from staining with Mitotracker to reveal mitochondria (Mito), and merged mitochondrial and Venus signals are shown. UVM11 is the Chlamydomonas strain used as the transformation host. Bars = 5 μm.