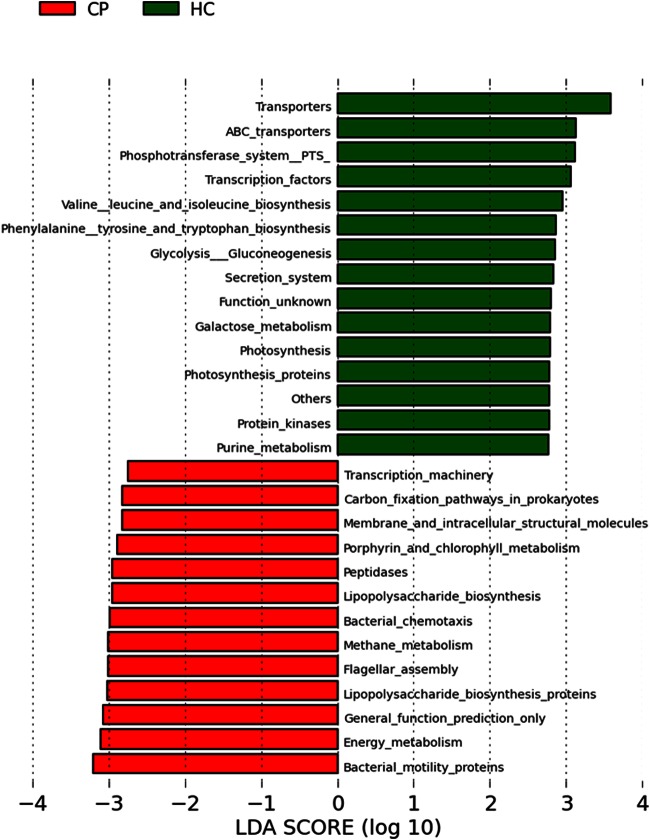
FIG 6.
Differentially abundant gene functions in subjects with chronic periodontitis and healthy controls. Functional categories of genes of the subgingival metagenome were predicted by using PICRUSt, and differentially abundant functions were then identified by using linear discriminant analysis (LDA) coupled with effect size measurements (LEfSe). Gene functions enriched in healthy sites are indicated with positive linear discriminant analysis scores, and functions differentially enriched in periodontitis sites are indicated with negative linear discriminant analysis scores. Only gene functions that have a linear discriminant analysis score threshold of 2.75 are shown.