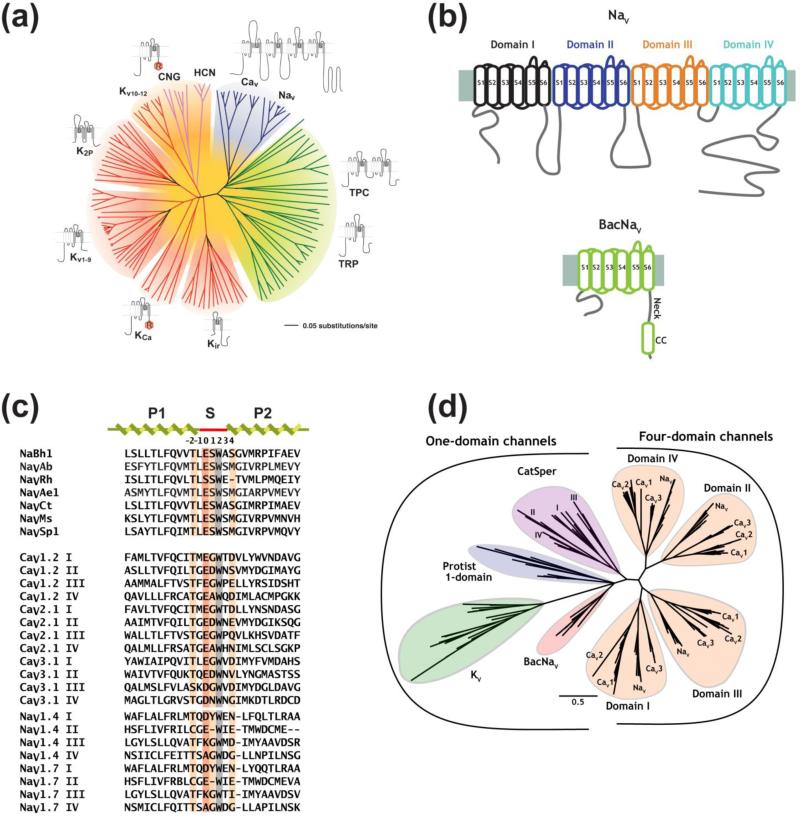
Figure 1. BacNaV topology and relationships to voltage-gated ion channel (VGIC) superfamily members.
(a) Unrooted tree showing the amino acid sequence relations of the minimal pore regions of VGIC superfamily members (modified from 10). Indicated subfamilies are (clockwise): voltage-gated calcium and sodium channels (CaV and NaV), two-pore (TPC) and transient receptor potential (TRP) channels, inwardly rectifying potassium channels (Kir), calcium-activated potassium channels (KCa), voltage-gated potassium channels (KV1–KV9), K2P channels, voltage-gated potassium channels from the EAG family (KV10–KV12), cyclic nucleotide gated channels (CNG), and hyperpolarization activated channels (HCN). “R” indicates identifiable regulatory domains. (b) Topology diagram comparing eukaryotic NaV (top) and BacNaV pore forming subunits. S1-S6 segments are labeled. Individual NaV six transmembrane repeats of are colored black, blue, orange, and teal. BacNaV neck and coiled coil (CC) domains are indicated. (c) Sequence alignment for selected BacNaVs selectivity filter and pore helices, mammalian CaV subtype exemplars, and mammalian NaV1.4 and NaV1.7. Selectivity filter numbering is indicated. Position (0) is highlighted in dark orange. Residues involved in selectivity are highlighted light orange. Grey highlights the the conserved Trp (+2) anchor position. (d) Unrooted tree showing a comparison based on the selectivity filter sequences for BacNaVs compared with KV channels, CatSper, Protist one-domain channels, and the individual domains of NaVs and CaVs (modified from 43).