Figure 1. A sub-group of germline genes remain refractory to activation by epigenetic modulating agents.
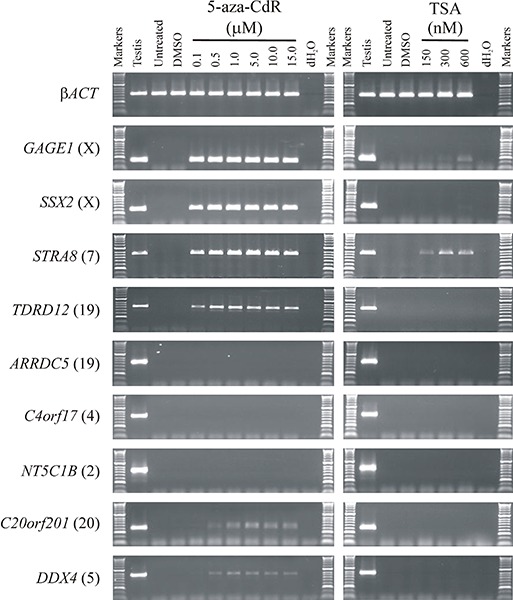
RT-PCR was used to analyse activation of a group of germline genes that are normally silenced in the cancer cell line HCT116 (an additional colorectal cell line gives similar results [see Supplementary Figure S1)]. Whilst a cohort of known and newly identified germline genes become activated at low doses of the demethylating agent 5-aza-CdR (GAGE1, SSX2, STRA8, TDRD12) and others become activated with slightly higher levels of 5-aza-CdR (C20orf201, DDX4), some remain tightly silenced, even at high concentrations of 5-aza-CdR (ARRDC5, C4orf17, NT5C1B) (left column). The histone deacetylase inhibitor trichostatin A (TSA) has little activating potential (other than for GAGE1 and STRA8, indicating the primary epigenetic regulation is mediated by DNA methylation (right column). Untreated and DMSO treated cells exhibit no activation of any of the genes analysed for expression activation. The chromosomal location of each gene is provided in parentheses to the right of the gene name. RT-PCR of βACT shows uniform sample quality and loading.
