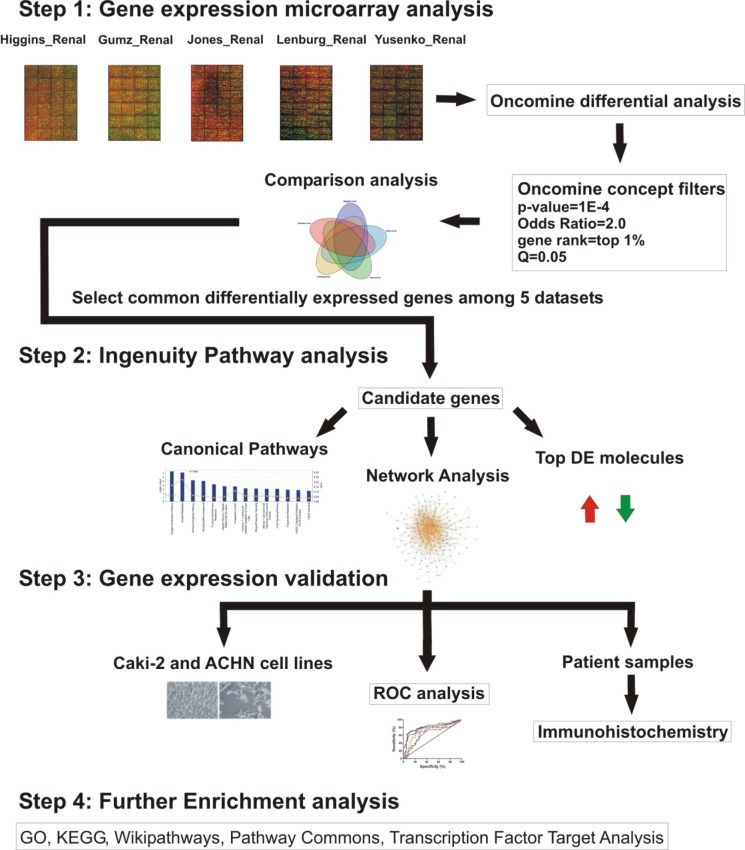
Figure 1. Workflow of the study.
Initially, five Oncomine microarray datasets were compared and the co-deregulated genes (co-DEGs) among them were retrieved. The co-DEGs were further enquired regarding their use as candidate markers for ccRCC. Next, the canonical pathways in which these co-DEGs are implicated were identified, as well as the networks that they form, and the top deregulated molecules among them. Following, validation of the deregulated expression levels of these genes was performed both in clear cell renal cell carcinoma cell lines, as well as in a cohort of ccRCC patients. Immunohistochemistry was performed in biopsies from the patient cohort for the top deregulated genes. ROC analysis was used to evaluate the discriminatory potential of the candidate biomarker genes. Further enrichment analysis was finally performed for the co-deregulated genes.