Figure 1.
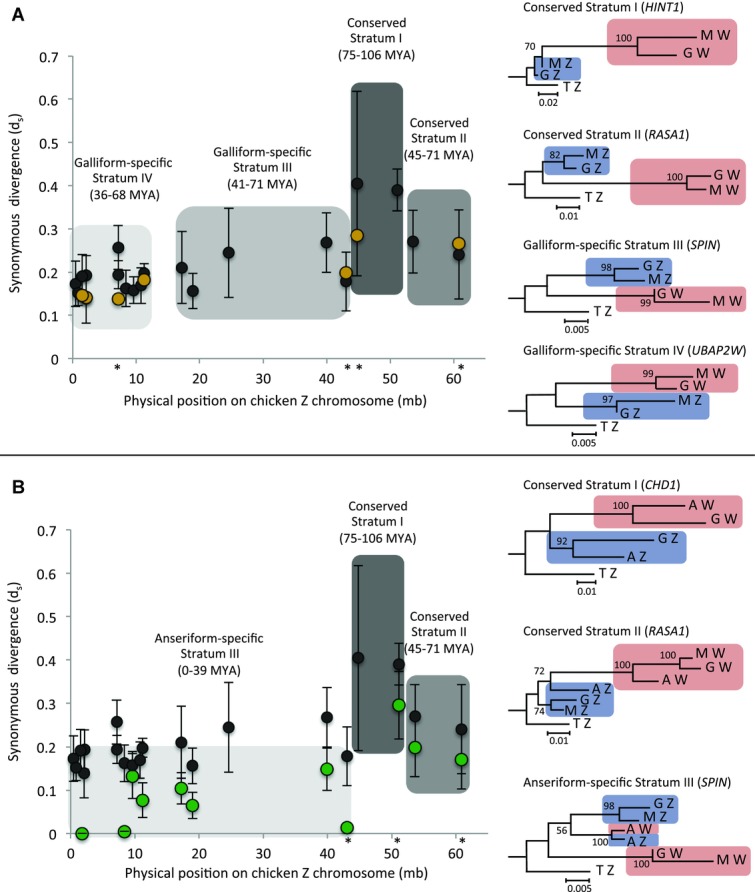
Dynamics of recombination suppression on the avian sex chromosomes. The distribution of synonymous (dS) divergence estimates between G. gallus (black), M. gallopavo (yellow, panel A) and A. platyrhynchos (green, panel B) gametologs, mapped against physical position of the G. gallus Z-linked ortholog. The 95% confidence intervals were calculated using standard errors. Bootstrap values were calculated using 1000 permutations. AZ, GZ, MZ, TZ corresponds to A. platyrhynchos, G. gallus, M. gallopavo, and T. guttata Z-linked genes. The physical Z-linked position of the loci for which gene trees are shown are indicated (*). Meleagris gallopavo dS estimates map closely onto the G. gallus strata boundaries, and maximum-likelihood gene trees for all gametologs cluster by sex chromosome, not species, indicating that four strata are conserved across the Galliformes (Fig. S2). Anas platyrhynchos dS estimates map closely onto the two oldest G. gallus strata boundaries, and maximum-likelihood gene trees for gametologs within these regions cluster by sex chromosome indicating that these strata are conserved across the Galloanserae. For the remaining gametologs, maximum-likelihood gene trees reveal that recombination was suppressed independently in the A. platyrhynchos lineage (Fig. S3). The dS estimates are highly heterogeneous and locus specific, indicating gradual divergence of gametologs with residual recombination across large tracts of the sex chromosomes.
