INTRODUCTION
According to the National Institutes of Health (NIH), bioinformatics includes “research, development, or application of computational tools and approaches for expanding the use of biological, medical, behavioral or health data, including those to acquire, store, organize, archive, analyze, or visualize such data” (2). With the increasing availability of biological information in online databases, building skills to use such databases is becoming ever more critical to those pursuing a biology career. Indeed, many claim that biology curricula cannot be considered relevant for the 21st century without inclusion of bioinformatics (1, 3, 5). However, not every campus has a resident bioinformaticist to help build curricula, and while there is an increasing number of bioinformatics exercises available on the Web, few of these have been formally evaluated. The two-part workshop described here included an initial 80-minute session with a guest speaker via Skype followed by an 80-minute session using exercises to demonstrate how bioinformatics tools could facilitate deeper understanding of human health and evolution for biology majors in an upper-division genetics class. Both sessions took place in a computer lab with Web access; no special software was required.
PROCEDURE
Introduction to basic local alignment search tool (BLAST)
The proposed outcomes of the first workshop were for students to gain an understanding of the nature and potential of bioinformatics as a field and as a set of tools, particularly in nucleotide sequence comparison (see Table 1). Students were asked to complete a pre-workshop survey (Appendix 1) before the first session. The survey asked them to define bioinformatics, answer six multiple-choice questions, and state their long-term career goal such that the perceived usefulness of bioinformatics within their desired careers could be ascertained.
TABLE 1.
Learning objectives of bioinformatics workshops.
| Learning Objectives | |
|---|---|
| Workshop 1 | Understand the kinds of questions that are answered in the field of bioinformatics. |
| Describe how BLAST assists in determining gene function. | |
| Workshop 2 | Determine the name and function of an unknown genetic sequence, as well as the relevance of that sequence to human health. |
| Describe how BLAST assists in finding homologous sequences between species, demonstrating the evolutionary heritage of human genes in other species. |
The first session was guided by an invited guest speaker, a bioinformatics researcher at an R1 institution, projected via Skype on a main screen. Guest speakers can be useful when expertise cannot be found on campus, and they have been recommended to help build campus-community relationships and improve student learning (4, 6). Having a guest speaker also allows the instructor to assist students at their computers while the guest is speaking and alert the guest to student questions. The guest speaker described the use of bioinformatics in his own research and then conducted a series of activities (Appendix 2) including introducing the website for the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), guiding students through finding a gene by name, obtaining the DNA sequence, and searching for similar sequences with the Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov/).
Application: Using BLAST to understand medicine and evolutionary biology
The learning outcomes of the second workshop were grounded in connecting bioinformatics to questions of evolution and medicine (see Table 1). Appendix 3 contains the activities completed in this second workshop, which were adapted from web-available exercises published by the East Bay Biotechnology Education Project (http://www.ebbep.com/docs/bioinformatics/introbioinformatics.pdf). These activities required students to determine the medical relevance of an unidentified disease-related sequence (Appendix 4), the homology of that sequence in other species, and the single nucleotide polymorphisms present in the sequence. Students could later be directed to background information on the diseases at Genetics Home Reference (http://ghr.nlm.nih.gov/). After the second workshop, students completed a post-workshop survey (Appendix 5). Survey data and statistics were analyzed using Microsoft Excel.
Tips and extensions
Provide instructions as a hyperlinked PDF or Word document so students can easily reach websites without having to type in web addresses manually.
Unidentified sequences can be adjusted for different student interests or class themes (such as genes involved with microbial pathogenesis, plant genes involved with agricultural adaptation, etc.).
CONCLUSION
Students’ perceptions of their own computer proficiency, the general usefulness of bioinformatics, and the usefulness of bioinformatics in their future careers did not change significantly upon completion of the workshop (Fig. 1, p > 0.05). However, students experienced significant gains when it came to their perceived understanding of the purpose of bioinformatics and knowledge of how to use bioinformatics tools (Fig. 2, p ≤ 0.05). In the pre- and post-workshop surveys, students were asked to define bioinformatics. In addition, students were given the same prompt to define bioinformatics on their final exam, more than six weeks after the second workshop session. A scoring rubric (Appendix 6) was developed to assess students’ understanding of bioinformatics based on its NIH definition (2). This assessment showed significant improvement in students’ understanding of bioinformatics after the two workshops, and indicated that those gains were retained six weeks later despite no further bioinformatics content being covered in the course (Table 2).
FIGURE 1.
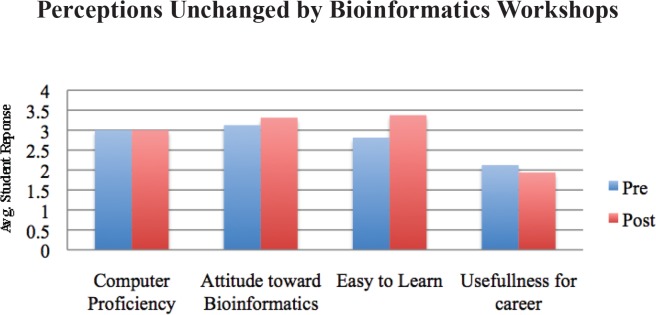
Perceptions unchanged by workshops: student responses to pre- and post-workshop survey where p > 0.05 in paired t-test.
FIGURE 2.
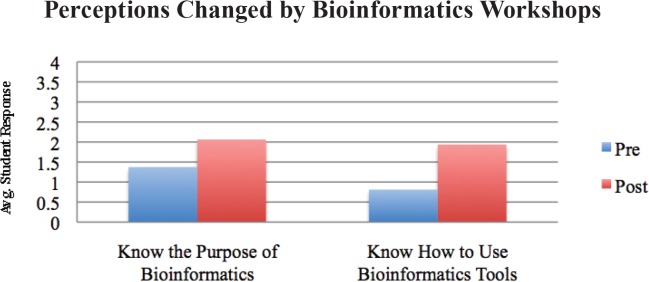
Perceptions changed by workshops: student responses to pre- and post-workshop survey where p < 0.05 in paired t-test.
TABLE 2.
Student gains in defining bioinformatics.
| Pre-Workshop | Post-Workshop | Final Exam |
|---|---|---|
| 2.00 | 2.50 | 2.45 |
Rubric found in Appendix 6, scores out of 4 possible points, n = 17, p = 0.0439 using a paired t-test.
SUPPLEMENTAL MATERIALS
Appendix 1: Pre-workshop survey
Appendix 2: Workshop session 1
Appendix 3: Workshop session 2
Appendix 4: Unidentified nucleotide sequences and instructor key
Appendix 5: Post-workshop survey
Appendix 6: Grading rubric for bioinformatics definition
Acknowledgments
I thank Richard Tillett, Bioinformatics Analyst at the University of Nevada, Reno, for being an excellent guest speaker and designing the first of the two workshops. The author declares that there are no conflicts of interest. No funding sources were used to collect data or create this report. The author takes full responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Supplemental materials available at http://jmbe.asm.org
REFERENCES
- 1.Bialek W, Botstein D. Introductory science and mathematics education for 21st century biologists. Science. 2004;303:788–790. doi: 10.1126/science.1095480. [DOI] [PubMed] [Google Scholar]
- 2.Huerta M, Downing G, Haseltine F, Seto B, Liu Y. NIH working definition of bioinformatics and computational biology. US National Institute of Health; Bethesda, MD: 2000. [Google Scholar]
- 3.Maloney M, Parker J, LeBlank M, Woodard CT, Glackin M, Hanrahan M. Bioinformatics and the undergraduate curriculum. CBE Life Sci Educ. 2010;9:172–174. doi: 10.1187/cbe.10-03-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mullins P. Using outside speakers in the classroom. APS Observer. 2001;14(8) [Google Scholar]
- 5.National Research Council Committee on Undergraduate Biology Education to Prepare Research Scientists for the 21st Century . BIO2010: Transforming undergraduate education for future research biologists. The National Academies Press; Washington, DC: 2003. [PubMed] [Google Scholar]
- 6.Wortmann GB. An invitation to learning: guest speakers in the classroom. Sci Teach. 1992;59:19–22. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 1: Pre-workshop survey
Appendix 2: Workshop session 1
Appendix 3: Workshop session 2
Appendix 4: Unidentified nucleotide sequences and instructor key
Appendix 5: Post-workshop survey
Appendix 6: Grading rubric for bioinformatics definition


