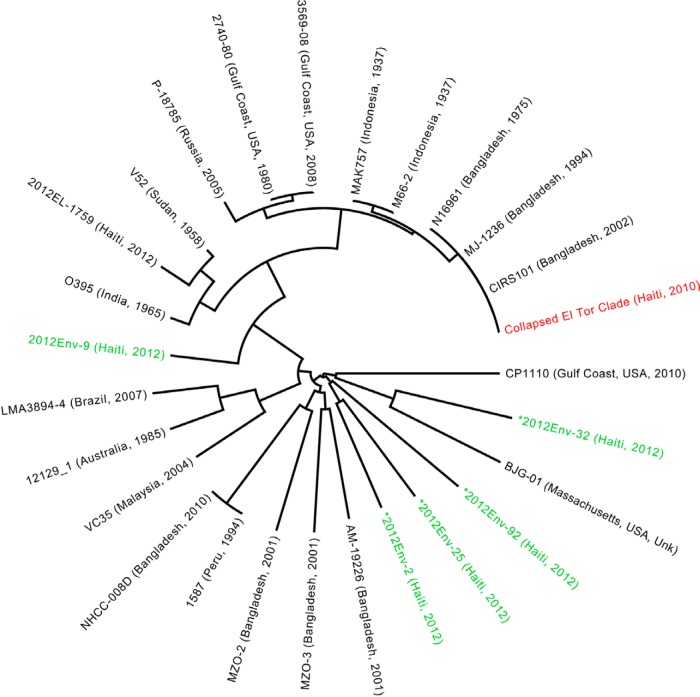
FIG 1 .
Maximum likelihood (ML) tree of V. cholerae environmental and clinical strains. The ML tree was inferred from ProgressiveMauve alignment of four environmentally sampled non-O1/O139 isolate sequences (designated by green coloring and marked with an asterisk), one environmentally sampled nontoxigenic O1 sequence, and 27 reference sequences. The 60 closely related toxigenic O1 biotype El Tor strains from our study are collapsed and represented by the 2010EL-1786 reference strain, which clusters with other members of the El Tor lineage. The four non-O1/O139 strains were dispersed throughout the global phylogeny, indicating a panmictic population of non-O1/O139 strains in the environment. The phylogeny was constructed with RAxML, using the GTR nucleotide substitution model and 1,000 bootstrap replicates. Bootstrap values for branches are not indicated but were all above 90%.