FIG 4 .
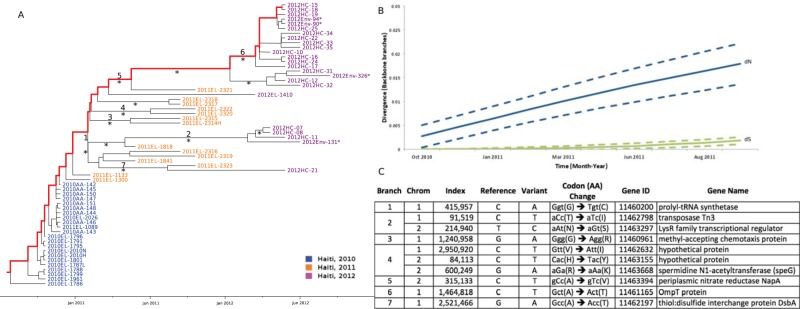
Bayesian MCC phylogeny and selection analysis along backbone paths. (A) Bayesian maximum clade credibility (MCC) tree of Haitian strains of V. cholerae with branch lengths scaled in time by enforcing a strict molecular clock. Strain labels are colored to indicate the time of sampling according to the legend in the figure. An asterisk along a branch marks subtending clades supported by a posterior probability of >0.75. An asterisk next to a node sequence name indicates an environmentally sampled isolate. One of the possible backbone paths, representing major lineages propagating over time from root node to most recently sampled sequence, is highlighted in red. Branch labels correspond to nonsynonymous mutations indicated in the table on panel C. (B) Mean nonsynonymous (dN) and synonymous (dS) divergence along backbone paths in a subsample of trees from the posterior distribution (see Materials and Methods) during the cholera epidemic in Haiti are represented by blue and green lines, respectively. Estimates 1 standard deviation above and below the mean dN or dS are represented by broken lines. The y axis represents the number of nucleotide (dN or dS) changes per (synonymous or nonsynonymous) SNP site. The x axis matches the time scale of the epidemic. Estimates were obtained by reconstructing and comparing ancestral sequences along V. cholerae trees sampled from the posterior distribution from the Bayesian analysis. (C) Inferred codon and amino acid changes in the V. cholerae genome (numbered according to the V. cholerae 2010EL-1786 reference strain) along internal branches of the O1 Haitian phylogeny. Branch numbers correspond to numbered branches in the MCC tree given in panel A.
