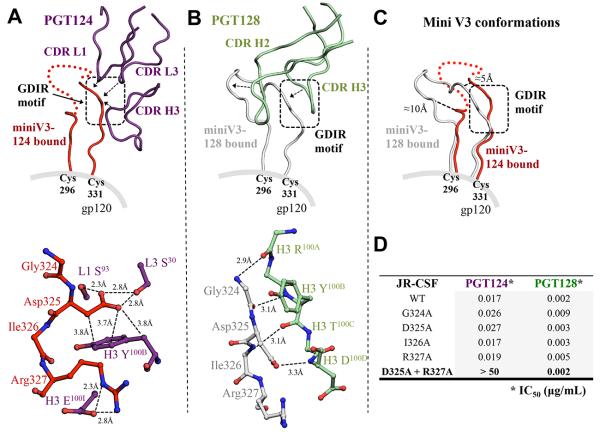
Figure 2. Interaction between PGT124 and the base of the V3 loop.
(A) The interaction between PGT124 CDR loops (purple ribbons) and the base of the V3 loop (red ribbon). Red dots represent residues at the tip of the mini V3 loop that are not visible in the electron density. The interaction with contacting residues is displayed in ball-and-stick representation below. Contacts between side chains of the 324GDIR327 motif on the V3 loop and the CDR loops of PGT124 are indicated with solid lines for van der Waals’ interactions and dotted lines for hydrogen bonds. (B) For comparison, the interaction between PGT128 CDR loops (pale green) and the base of the V3 (gray) (PDB 3TYG) for comparison with (A). The PGT128 antibody-antigen interactions with the V3 base are all H-bonds of the Fab main chain with V3 main chain. (C) The V3 loop structures bound to PGT124 or PGT128 are superimposed for comparison. (D) The effect on the neutralization activity of PGT124 and PGT128 by single and double mutations of the V3 loop GDIR motif. Values are shown as IC50 in μg/mL (see also Figures S3, S4 and S5).