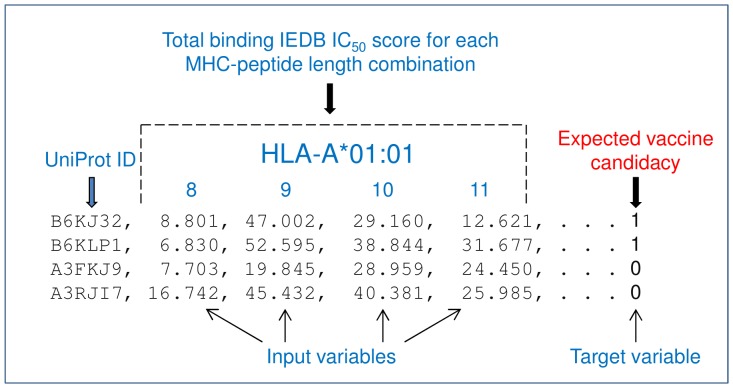
Figure 3. Example file format of training dataset used in machine learning.
There is one protein per line that consists of the total binding affinity score for each peptide-MHC length combination e.g. 304 combinations for 76 common MHC I alleles (MHC I binds to peptides, typically eight to eleven amino acid residues in length. Therefore, 76 alleles * 4 peptide lengths = 304 combinations). Binding affinity score = an IEDB IC50 (nM) score <5000. Each score is weighted by the length of the protein. The scores represent input variables or predictors. The last column is a 1 or 0 that indicates an expected ‘YES’ or ‘NO’ vaccine candidacy and represents the target variable. This expectation is based on the subcellular location annotation associated with the protein in UniProtKB (secreted or membrane-associated = 1, internal location = 0).