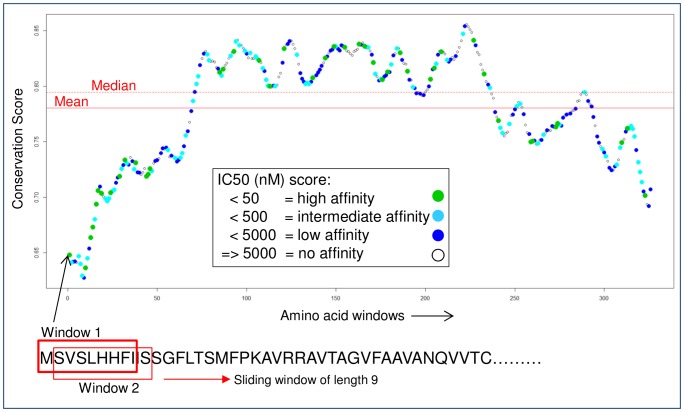
Figure 4. Plot of conservation scores computed for binding peptides along a protein (UniProtKB ID: P13664).
Each circle represents the amino acid conservation score computed at a sliding window. The window is of length 9 and slides one residue at a time. The colour of the circle represents binding affinities against 76 common MHC alleles computed at each window. A window (i.e. a peptide) can theoretically bind to all 76 alleles and colours are therefore plotted in a set order: no, low, intermediate, and high affinity. For example, a dark blue circle for low affinity indicates there are no intermediate or high affinity peptides at the window; however, a green circle for high affinity provides no indication of other affinities at the same window. Mean conservation = 0.7805; median conservation = 0.7946. For protein P13664 (Major surface antigen p30) 54.6% high, 56% intermediate, and 55.9% low binders have conservation scores below the mean. The study shows that vaccine candidates are significantly more likely to have either a greater number of less conserved peptides or a lower total conservation score than non-vaccine candidates.