Abstract
Background
Alternative splicing (AS) is a key regulatory mechanism in protein synthesis and proteome diversity. In this study, we identified alternative splicing events in four pairs of cervical squamous cell carcinoma (CSCC) and adjacent nontumor tissues using RNA sequencing.
Methods
The transcripts of the four paired samples were thoroughly analyzed by RNA sequencing. SpliceMap software was used to detect the splicing junctions. Kyoto Encyclopedia of Genes and Genomes pathway analysis was conducted to detect the alternative spliced genes-related signal pathways. The alternative spliced genes were validated by reverse transcription-polymerase chain reaction (RT-PCR).
Results
There were 35 common alternative spliced genes in the four CSCC samples; they were novel and CSCC specific. Sixteen pathways were significantly enriched (P<0.05). One novel 5′AS site in the KLHDC7B gene, encoding kelch domain-containing 7B, and an exon-skipping site in the SYCP2 gene, encoding synaptonemal complex 2, were validated by RT-PCR. The KLHDC7B gene with 5′AS was found in 67.5% (27/40) of CSCC samples and was significantly related with cellular differentiation and tumor size. The exon-skipping site of the SYCP2 gene was found in 35.0% (14/40) of CSCC samples and was significantly related with depth of cervical invasion.
Conclusion
The KLHDC7B and the SYCP2 genes with alternative spliced events might be involved in the development and progression of CSCC and could be used as biomarkers in the diagnosis and prognosis of CSCC.
Keywords: cervical squamous carcinomas, alternative splicing events, RNA sequencing
Introduction
Alternative splicing (AS) is a biological process by which different exons are joined together to generate a series of mRNA isoforms from a single primary transcript. Nearly 90% of human multiple-exon genes are alternatively spliced, and AS is a common mechanism for generating both different transcription products and protein diversity in higher eukaryotic cells.1 The roles of AS in human diseases, especially in cancer, have been widely studied.2 Tumor formation might be due to the imbalanced expressions of either the splicing variants or the incorrect isoforms.3 Many oncogenes and tumor suppressor genes, such as BRCA1/24 and p53,5 are alternatively spliced in cancer cells. The cancer-specific isoforms induce the phenotypic transformation of cancer cells.6,7 Transcript sequencing has indicated that the gene mutations associated with cancer-specific AS events could be potentially used as valuable biomarkers in the diagnosis and therapy of cancer.8
Cervical cancer is the third most commonly diagnosed cancer and the fourth leading cause of cancer deaths among women around the world.9 Cervical cancer comprises 80% of squamous cell carcinoma (CSCC).10 The etiology of cervical cancer is absolutely related to persistent infection by human papillomavirus (HPV).11 The carcinogenesis due to HPV depends on the activities of viral oncoproteins E5, E6, and E7, which inhibit various cellular targets, including the tumor suppressor proteins p53, pRb, p21, and p27, as well as disrupting critical cellular processes, including cell cycle, apoptosis, and malignant transformation of cervical basal cells.12 In high-risk HPV types, transcription is initiated at the early promoter located in the E6 open reading frame (ORF) and the late promoter in the E7 ORF of HPV. All viral genes are transcribed to many polycistronic RNAs with two or more ORFs, which then undergo further processing, including AS and polyadenylation.13 For HPV16, at least 13 different mRNAs with the capacity to encode capsid proteins are produced by AS.14
In this study, we detected the CSCC-specific AS events by comparing the global transcriptional changes of CSCC to that of the adjacent nontumor tissues (ATN) through RNA sequencing. This study aims to advance our understanding of CSCC.
Patients and methods
Tissue specimens
Forty paired fresh-frozen tissue samples (CSCC and ATN) were collected from patients receiving radical hysterectomy for CSCC during the period of January 2012 to August 2013 (Peking Union Medical College Hospital, People’s Republic of China). Diagnosis of all cases was histologically confirmed by two independent pathologists, and all tumor tissues were assessed by hematoxylin–eosin (HE) staining, and only those tissues with percentage of tumor cells more than 90% were used. Four paired samples were randomly selected for RNA sequencing from among these cases. Informed consent from each patient was obtained. The procedures have been approved by the ethics review committee of Peking Union Medical College Hospital and are in accordance with the Helsinki Declaration of 1975.
Raw read filtering
The complementary DNA (cDNA) library of the four paired samples was constructed and sequenced. The raw RNA-sequencing data were filtered according to the following criteria: 1) reads containing sequencing adaptors were removed; 2) nucleotides with a quality score <20 were removed; 3) reads with more than 8% nitrogenous bases were removed. All subsequent analyses were based on clean reads.15
Detection of AS
SpliceMap was used to detect splicing junctions and different types of AS events, including exon skipping, mutually exclusive exons, intron retention, 5′AS, and 3′AS in CSCC and ATN tissues.16 The read was separated into segments. Each segment was mapped to the human genome with Bowtie software.17 Then all of the segments were pieced together to determine the locations of exons and possible junctions. We filtered the splice junctions originally detected according to two criteria: quality of the alignment and coverage of splice junction. The AS events presented only in the CSCC or ATN samples were detected.
Kyoto Encyclopedia of Genes and Genomes pathway analysis
The unique lists of CSCC-specific AS genes were submitted to the Web-based functional annotation tool, which is known as the Database for Annotation, Visualization and Integrated Discovery v6.7.18 The false discovery rate (FDR) was set at 5%, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was conducted for functional annotation categories.
Validation of AS genes by reverse transcriptase–polymerase chain reaction
Total RNA was extracted from frozen CSCC and ATN samples, and cDNA was synthesized from 5 μg of RNA (Invitrogen). To validate AS genes, reverse transcriptase-polymerase chain reaction (RT-PCR) was performed using AS gene-specific PCR primers in CSCC and ATN tissues, with cycling parameters: 95°C for 10 minutes; followed by 35 cycles each of 95°C for 45 seconds, annealing at 55°C or 60°C for 45 seconds, and extension at 72°C for 45 seconds; and a final extension at 72°C for 10 minutes. Electrophoresis on 1.5% agarose gels was carried out for PCR products. The primers were as follows: KLHDC7B (kelch domain-containing 7B) sense primer: 5′-AGGTGAGGCTCAGACAAGA-3′, antisense primer: 5′-GAGATGGTGGGAGAATGG-3′; SYCP2 (synaptonemal complex 2) sense primer: 5′-GATTACGGTGTCAGGAGG-3′, antisense primer: 5′-CTGGGAGATAAGTCAAGG-3′; GAPDH (glyceraldehyde-3-phosphate dehydrogenase) sense primer: 5′-GTCAAGGCTGAGAACGGGAA-3′, antisense primer: 5′-AAATGAGCCCCAGCCTTCTC-3′. GAPDH was used as the reference gene.
Correlation between AS genes and the clinicopathologic characteristics of CSCC patients was tested by the chi-square test. Fisher’s exact text was used when theoretical frequency was <5.0. Statistical significance was assumed as P<0.05. Statistical analysis was performed using the SPSS 13.0 statistical software.
Results
Characterization of RNA sequences
Four paired samples were collected for RNA sequencing. We obtained 2.0 million, 2.6 million, 0.8 million, and 2.2 million paired reads from the four CSCC samples, respectively. Similarly, 2.8 million, 5.4 million, 2.4 million, and 1.6 million paired reads were obtained from the ATN samples accordingly. The mapped ratio ranged from 63.1% to 73.2%.
Detection of CSCC-specific AS genes
SpliceMap was used to detect splice junctions. We compared the CSCC transcripts and ATN transcripts with the reference genome (Table 1). There were 17,462, 25,101, 9,034, and 25,925 newly detected AS events in the CSCC tissues and 17,161, 25,101, 19,901, and 13,279 newly detected AS events in the ATN tissues from the four paired samples. We screened out the AS events with more than one mapped reads; thus, 307, 555, 86, and 603 specific AS events were present in the four CSCC tissues, respectively. There were 35 common AS genes among the four CSCC tissues. One novel junction with a 5′AS site in the KLHDC7B gene was supported by 1, 3, 18, and 22 reads in the four CSCC tissues, and an exon-skipping site in the SYCP2 gene was supported by 2, 3, 12, and 13 reads in the four CSCC tissues (Table S1).
Table 1.
Newly detected AS events in CSCC and ATN samples
| Sample | AS events in sample transcripts | AS events in reference transcripts | Newly detected AS events |
|---|---|---|---|
| LHR-CSCC | 147,341 | 129,879 | 17,462 |
| LHR-ATN | 142,583 | 125,422 | 17,161 |
| LYM-CSCC | 166,558 | 141,457 | 25,101 |
| LYM-ATN | 194,893 | 154,596 | 40,297 |
| ZTF-CSCC | 122,182 | 113,148 | 9,034 |
| ZTF-ATN | 153,975 | 134,074 | 19,901 |
| WSN-CSCC | 170,865 | 144,940 | 25,925 |
| WSN-ATN | 116,545 | 103,266 | 13,279 |
Abbreviations: AS, alternative splicing; CSCC, cervical squamous cell carcinoma; ATN, adjacent nontumor tissues.
KEGG pathway analysis
KEGG pathway analysis was used to identify AS gene-related significantly enriched pathways. In total, 16 pathways were significantly enriched (P<0.05), including metabolic pathways, endocytosis, and the Ras signaling pathway, and all of these pathways were specific for CSCC (Table 2).
Table 2.
Signal pathways related with CSCC-specific AS events (P<0.05)
| KEGG entry | Signal pathway | Gene number |
|---|---|---|
| Ko05144 | Malaria | 6 |
| Ko04270 | Vascular smooth muscle contraction | 7 |
| Ko04962 | Vasopressin-regulated water reabsorption | 4 |
| Ko01100 | Metabolic pathways | 23 |
| Ko05152 | Tuberculosis | 12 |
| Ko04670 | Leukocyte transendothelial migration | 5 |
| Ko04145 | Phagosome | 6 |
| Ko05016 | Huntington’s disease | 8 |
| Ko04014 | Ras signaling pathway | 10 |
| Ko04144 | Endocytosis | 12 |
| Ko04514 | Cell adhesion molecules | 4 |
| Ko04120 | Ubiquitin-mediated proteolysis | 6 |
| Ko04142 | Lysosome | 5 |
| Ko04530 | Tight junction | 6 |
| Ko00563 | GPI-anchor biosynthesis | 5 |
| Ko05146 | Amebiasis | 4 |
Abbreviations: AS, alternative splicing; CSCC, cervical squamous cell carcinoma; GPI, glycosylphosphatidylinositol; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Validation of AS genes
KLHDC7B and SYCP2 genes were chosen as the candidate genes according to the number of mapped reads in the four pairs of samples. One novel junction with a 5′AS site in the KLHDC7B gene (Figure 1) and an exon-skipping site in the SYCP2 gene were found (Figure 2). The AS events in the KLHDC7B (Figure 3A) and the SYCP2 genes (Figure 3B) were CSCC specific and were validated by RT-PCR. In total, the KLHDC7B gene with 5′AS was found in 67.5% (27/40) of CSCC samples and was positively related with cellular differentiation and tumor size (Table 3). The exon-skipping site of the SYCP2 gene was found in 35.0% (14/40) of CSCC samples and was positively related with the depth of cervical invasion (Table 3).
Figure 1.
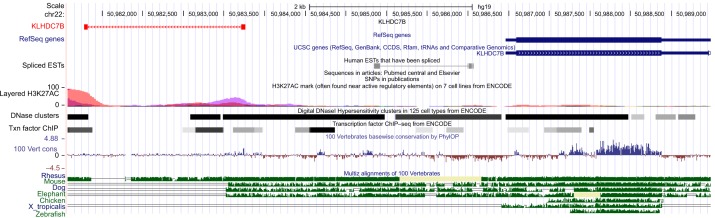
Analysis of novel AS junctions in KLHDC7B gene on the UCSC genome browser of the human genome built via custom track.
Notes: The novel splice junctions of the genes are indicated at the top followed by UCSC genes and mammalian conservation of the corresponding regions. The red labels indicate the 5′AS junction of the KLHDC7B gene.
Abbreviations: AS, alternative splicing; EST, expressed sequence tags; ChIP, chromatin immunoprecipitation; KLHDC7B, kelch domain-containing 7B; SNPs, single-nucleotide polymorphisms; UCSC, University of California, Santa Cruz.
Figure 2.
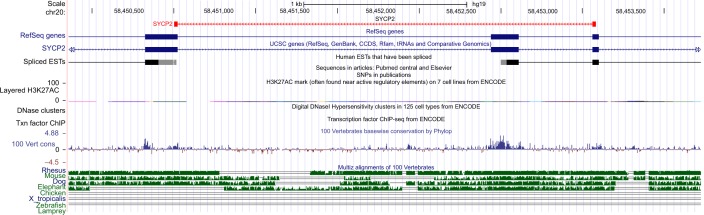
Analysis of novel AS junction in SYCP2 gene on UCSC genome browser of the human genome built via custom track.
Notes: The novel splice junctions of genes are indicated at the top followed by UCSC genes and mammalian conservation of the corresponding regions. The red labels indicated the exon-skipping splice junction of the SYCP2 gene.
Abbreviations: AS, alternative splicing; EST, expressed sequence tags; ChIP, chromatin immunoprecipitation; SNPs, single-nucleotide polymorphisms; SYCP2, synaptonemal complex 2; UCSC, University of California, Santa Cruz.
Figure 3.
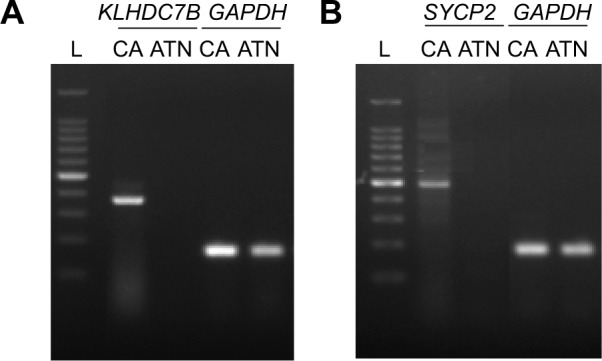
The AS events of the KLHDC7B and SYCP2 genes were validated by RT-PCR.
Notes: (A) The 5′AS junction of KLHDC7B gene was CSCC specific. (B) The exon skipping of SYCP2 gene was CSCC specific. GAPDH was used as the reference gene. L: O’RangeRuler 100-bp DNA ladder.
Abbreviations: AS, alternative splicing; ATN, adjacent nontumor tissues; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; KLHDC7B, kelch domain-containing 7B; RT-PCR, reverse transcriptase-polymerase chain reaction; SYCP2, synaptonemal complex 2; CA, CSCC; CSCC, cervical squamous cell carcinoma.
Table 3.
Correlation between AS genes and the clinicopathologic characteristics of CSCC patients
| Variable feature | n |
KLHDC7B
|
P-value |
SYCP2
|
P-value | ||
|---|---|---|---|---|---|---|---|
| + | − | + | − | ||||
| Age | |||||||
| ≥40 | 24 | 17 | 7 | 0.304 | 6 | 18 | 0.104 |
| <40 | 16 | 10 | 6 | 8 | 8 | ||
| Differentiation | |||||||
| Good | 17 | 10 | 7 | 0.012 | 5 | 12 | 0.058 |
| Moderate | 16 | 12 | 4 | 7 | 9 | ||
| Poor | 7 | 5 | 2 | 2 | 5 | ||
| Tumor size (cm) | |||||||
| ≥4.0 | 18 | 16 | 2 | 0.016 | 9 | 9 | 0.072 |
| <4.0 | 22 | 11 | 11 | 5 | 17 | ||
| Lymphatic metastasis | |||||||
| Yes | 8 | 5 | 3 | 0.736 | 3 | 5 | 0.868 |
| No | 32 | 22 | 10 | 11 | 21 | ||
| Cervical invasion | |||||||
| ≥1/2 | 12 | 8 | 4 | 0.94 | 8 | 4 | 0.006 |
| <1/2 | 28 | 19 | 9 | 6 | 22 | ||
| FIGO staging | |||||||
| Ia | 6 | 4 | 2 | 0.518 | 1 | 5 | 0.495 |
| Ib | 27 | 17 | 10 | 11 | 16 | ||
| IIa | 7 | 6 | 1 | 2 | 5 | ||
Note: Fisher’s exact text was used when theoretical frequency was <5.
Abbreviations: AS, alternative splicing; CSCC, cervical squamous cell carcinoma; FIGO, International Federation of Gynecology and Obstetrics; KLHDC7B, kelch domain-containing 7B; SYCP2, synaptonemal complex 2.
Discussion
AS is crucial in normal development programs and its dysregulation is related to tumorigenesis.19,20 AS events are involved in cell cycle, metabolism, tumor suppression, and various cell signaling pathways.21 The cancer-specific AS events can lead to the activation of oncogenes and cancer-related pathways, as well as inactivation of tumor suppressors.22 In this study, a total of 35 novel CSCC-specific AS genes and 16 significant pathways were identified.
Metabolic pathways, endocytosis, and the Ras signaling pathway were the three most important pathways detected by KEGG pathway analysis. It has been reported that metabolic alternations contribute to the development of cancer.23 The major metabolic pathways, such as glycolysis and oxidative phosphorylation, are altered in cancer cells to meet the bioenergetic and biosynthetic demands associated with tumor growth.24,25 The process of endocytosis and endocytic proteins are involved in the regulation of cell cycle, mitosis, and apoptosis in cancer cells.26,27 The AS isoforms of Ras were able to activate the MAP kinase signaling pathway and to induce tumor formation in nude mice.28 The Ras/Raf/MAPK cascade can be activated by the epidermal growth factor receptor (EGFR/ErbB1), a member of the ErbB receptor tyrosine kinase family, which is frequently mutated and overexpressed in different human cancers, including glioma, non-small cell lung carcinoma, ovarian carcinoma, and prostate carcinoma.29 This research demonstrated that the AS genes in these signal pathways might participate in the progress of CSCC.
On the basis of the RNA-sequencing analysis, we confirmed the AS events in KLHDC7B and SYCP2 genes in CSCC tissues by RT-PCR for the first time. The KLHDC7B mRNA is increased under several biological conditions mainly due to infection. KLHDC7B expression was increased during acute HCV infection and was induced by interferon gamma, TNF-α, and IL-4. The KLHDC7B gene regulates and facilitates HCV replication in hepatocytes.30,31 In the study of Kim et al the KLHDC7B gene containing a kelch domain was identified as a candidate novel epigenetic marker that was hypermethylated and upregulated in breast cancer; the methylation level of the 14CpG sites at the promoter region of the gene was higher in cancer tissues and cultured breast cell lines.32 KLHDC7B was upregulated in vulvar intraepithelial neoplasia due to HPV infection.33 We speculated that the spliced KLHDC7B gene might bind to HPV oncoproteins to promote CSCC progression.
SYCP2 is a proteinaceous structure that links homologous chromosomes during the prophase of meiosis. The protein encoded by this gene is a major component of the synaptonemal complex and may bind DNA at scaffold attachment regions. It had been reported that SYCP2 was upregulated in Caski and SiHa cells and is associated with invasive cervical cancer.34,35
The expression of the KLHDC7B gene with 5′AS was positively related with cellular differentiation and tumor size. The SYCP2 gene with exon skipping was positively related with depth of cervical invasion. The AS events in KLHDC7B and SYCP2 genes might generate new transcripts or regulatory proteins to promote the progress of CSCC, and KLHDC7B and SYCP2 genes with the novel AS events could be used as potential biomarkers in diagnosis and therapy of CSCC patients. However, the mechanisms of action of the two genes with AS events in the carcinogenesis of CSCC need further investigation.
Supplementary material
Table S1.
The alternative splicing junctions of the four paired CSCC and ATN tissues
| Junction | Strand | Official gene name | LHR | LYM | ZTF | WSN |
|---|---|---|---|---|---|---|
| chr9:100767385–100773605 | + | ANP32B | (1/0) | (2/0) | (1/0) | (3/0) |
| chr1:111985966–111986338 | − | WDR77 | (1/0) | (2/0) | (1/0) | (2/0) |
| chr1:156015471–156018341 | − | UBQLN4 | (1/0) | (1/0) | (1/0) | (1/0) |
| chr1:156563314–156563723 | + | APOA1BP | (1/0) | (2/0) | (1/0) | (2/0) |
| chr1:160959034–160967990 | + | F11r | (1/0) | (8/0) | (2/0) | (3/0) |
| chr1:1770627–1784640 | − | GNB1 | (1/0) | (2/0) | (1/0) | (5/0) |
| chr1:241799325–241803233 | − | OPN3 | (2/0) | (1/0) | (1/0) | (6/0) |
| chr10:103769156–103771529 | − | c10orf76 | (2/0) | (1/0) | (2/0) | (1/0) |
| chr10:134143939–134145035 | − | LRRC27 | (1/0) | (1/0) | (2/0) | (2/0) |
| chr12:100574444–100589409 | − | microRNA 1827 | (1/0) | (1/0) | (2/0) | (1/0) |
| chr13:103436812–103440233 | − | Kdelc1 | (1/0) | (2/0) | (1/0) | (1/0) |
| chr13:77703315–77713380 | − | MYCBP2 | (1/0) | (2/0) | (1/0) | (4/0) |
| chr13:79889075–79893005 | − | RBM26 | (2/0) | (1/0) | (1/0) | (1/0) |
| chr14:103589770–103592696 | + | TNFAIP2 | (3/0) | (4/0) | (2/0) | (11/0) |
| chr15:30939867–30951018 | + | ARHGAP11B | (1/0) | (2/0) | (1/0) | (2/0) |
| chr15:68489916–68521889 | − | calml4 | (1/0) | (1/0) | (1/0) | (1/0) |
| chr15:93444004–93467600 | + | CHD2 | (2/0) | (2/0) | (1/0) | (2/0) |
| chr16:55463342–55478074 | − | MMP2 | (1/0) | (1/0) | (2/0) | (3/0) |
| chr19:13228895–13246063 | + | NACC1 | (1/0) | (1/0) | (1/0) | (1/0) |
| chr19:48653417–48657178 | − | lig1 | (1/3) | (2/0) | (1/0) | (1/0) |
| chr20:32224464–32228199 | + | CBFA2T2 | (2/0) | (2/0) | (1/0) | (1/0) |
| chr20:57484584–57485055 | + | Gnas | (1/0) | (1/0) | (1/0) | (1/0) |
| chr20:58450474–58453112 | − | sycp2 | (13/0) | (12/0) | (2/0) | (3/0) |
| chr22:24179991–24180800 | − | Derl3 | (2/0) | (1/0) | (1/0) | (4/0) |
| chr22:50981278–50983265 | − | KLHDC7B | (3/0) | (18/0) | (1/0) | (22/0) |
| chr4:119459113–119461423 | + | CEP170P1 | (0/2) | (0/3) | (0/2) | (0/5) |
| chr4:184255479–184255672 | − | CLDN24 | (1/0) | (2/0) | (1/0) | (1/0) |
| chr5:33466262–33467025 | + | Tars | (1/0) | (4/0) | (2/0) | (1/0) |
| chr6:52128956–52129242 | − | MCM3 | (4/0) | (3/0) | (2/0) | (3/0) |
| chr7:6066538–6068295 | − | Eif2ak1 | (1/0) | (1/0) | (1/0) | (1/0) |
| chr8:145666203–145667469 | − | TONSL | (1/0) | (1/0) | (1/0) | (1/0) |
| chr8:74872003–74884326 | − | TCEB1 | (2/0) | (1/0) | (2/0) | (1/1) |
| chr9:5436644–5438262 | − | PLGRKT | (1/0) | (1/0) | (2/0) | (1/0) |
| chr9:99086401–99122485 | − | SLC35D2 | (1/0) | (1/0) | (1/0) | (1/0) |
Abbreviations: ATN, adjacent nontumor tissues; CSCC, cervical squamous cell carcinoma.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (81472446).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med. 2012;18(8):472–482. doi: 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonomi S, Gallo S, Catillo M, Pignataro D, Biamonti G, Ghigna C. Oncogenic alternative splicing switches: role in cancer progression and prospects for therapy. Int J Cell Biol. 2013;2013:962038. doi: 10.1155/2013/962038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mucaki EJ, Ainsworth P, Rogan PK. Comprehensive prediction of mRNA splicing effects of BRCA1 and BRCA2 variants. Hum Mutat. 2011;32(7):735–742. doi: 10.1002/humu.21513. [DOI] [PubMed] [Google Scholar]
- 5.Wei J, Zaika E, Zaika A. p53 family: role of protein isoforms in human cancer. J Nucleic Acids. 2012;2012:687359. doi: 10.1155/2012/687359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Lee W, Jiang Z, et al. Genome and transcriptome sequencing of lung cancers reveal diverse mutational and splicing events. Genome Res. 2012;22(12):2315–2327. doi: 10.1101/gr.140988.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venables JP, Klinck R, Koh C, et al. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol. 2009;16(6):670–676. doi: 10.1038/nsmb.1608. [DOI] [PubMed] [Google Scholar]
- 8.Li W, Dai C, Kang S, Zhou XJ. Integrative analysis of many RNA-seq datasets to study alternative splicing. Methods. 2014;67(3):313–324. doi: 10.1016/j.ymeth.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 10.Mathew A, George PS. Trends in incidence and mortality rates of squamous cell carcinoma and adenocarcinoma of cervix-worldwide. Asian Pac J Cancer Prev. 2009;10(4):645–650. [PubMed] [Google Scholar]
- 11.Bernard HU, Burk RD, Chen Z, van Doorslaer K, zur Hausen H, de Villiers EM. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401(1):70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLaughlin-Drubin ME, Meyers J, Munger K. Cancer associated human papillomaviruses. Curr Opin Virol. 2012;2(4):459–466. doi: 10.1016/j.coviro.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham SV. Human papillomavirus: gene expression, regulation and prospects for novel diagnostic methods and antiviral therapies. Future Microbiol. 2010;5(10):1493–1506. doi: 10.2217/fmb.10.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milligan SG, Veerapraditsin T, Ahamat B, Mole S, Graham SV. Analysis of novel human papillomavirus type 16 late mRNAs in differentiated W12 cervical epithelial cells. Virology. 2007;360(1):172–181. doi: 10.1016/j.virol.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Y, Wang X, Wu F, et al. Transcriptome profiling of the cancer, adjacent non-tumor and distant normal tissues from a colorectal cancer patient by deep sequencing. PLoS One. 2012;7(8):e41001. doi: 10.1371/journal.pone.0041001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Au KF, Jiang H, Lin L, Xiang Y, Wong WH. Detection of splice junctions from paired-end RNA-seq data by SpliceMap. Nucleic Acids Res. 2010;38(14):4570–4578. doi: 10.1093/nar/gkq211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferragina P, Manzini G. An experimental study of an opportun-istic index; Proceedings of the twelfth annual ACM-SIAM symposium on discrete algorithms; Washington, DC: Society for industrial and applied mathematics; 2001. pp. 269–278. [Google Scholar]
- 18.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.Barash Y, Calarco JA, Gao W, et al. Deciphering the splicing code. Nature. 2010;465(7249):53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 20.Shah TM, Patel AK, Bhatt VD, et al. The landscape of alternative splicing in buccal mucosa squamous cell carcinoma. Oral Oncol. 2013;49(6):604–610. doi: 10.1016/j.oraloncology.2013.03.431. [DOI] [PubMed] [Google Scholar]
- 21.Biamonti G, Catillo M, Pignataro D, Montecucco A, Ghigna C. The alternative splicing side of cancer. Semin Cell Dev Biol. 2014;32C:30–36. doi: 10.1016/j.semcdb.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 22.Biamonti G, Bonomi S, Gallo S, Ghigna C. Making alternative splicing decisions during epithelial-to-mesenchymal transition (EMT) Cell Mol Life Sci. 2012;69(15):2515–2526. doi: 10.1007/s00018-012-0931-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 25.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boucrot E, Kirchhausen T. Endosomal recycling controls plasma membrane area during mitosis. Proc Natl Acad Sci U S A. 2007;104(19):7939–7944. doi: 10.1073/pnas.0702511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehtonen S, Shah M, Nielsen R, et al. The endocytic adaptor protein ARH associates with motor and centrosomal proteins and is involved in centrosome assembly and cytokinesis. Mol Biol Cell. 2008;19(7):2949–2961. doi: 10.1091/mbc.E07-05-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baitei EY, Zou M, Al-Mohanna F, et al. Aberrant BRAF splicing as an alternative mechanism for oncogenic B-Raf activation in thyroid carcinoma. J Pathol. 2009;217(5):707–715. doi: 10.1002/path.2496. [DOI] [PubMed] [Google Scholar]
- 29.Rowinsky EK. The erbB family: targets for therapeutic development against cancer and therapeutic strategies using monoclonal antibodies and tyrosine kinase inhibitors. Annu Rev Med. 2004;55:433–457. doi: 10.1146/annurev.med.55.091902.104433. [DOI] [PubMed] [Google Scholar]
- 30.Sana TR, Janatpour MJ, Sathe M, McEvoy LM, McClanahan TK. Microarray analysis of primary endothelial cells challenged with different inflammatory and immune cytokines. Cytokine. 2005;29(6):256–269. doi: 10.1016/j.cyto.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 31.Papic N, Maxwell CI, Delker DA, Liu S, Heale BS, Hagedorn CH. RNA-sequencing analysis of 5′ capped RNAs identifies many new differentially expressed genes in acute hepatitis C virus infection. Viruses. 2012;4(4):581–612. doi: 10.3390/v4040581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim TW, Kim YJ, Lee HJ, Min SY, Kang HS, Kim SJ. Hs.137007 is a novel epigenetic marker hypermethylated and up-regulated in breast cancer. Int J Oncol. 2010;36(5):1105–1111. doi: 10.3892/ijo_00000592. [DOI] [PubMed] [Google Scholar]
- 33.Santegoets LA, Seters MV, Helmerhorst TJ, et al. HPV related VIN: highly proliferative and diminished responsiveness to extracellular signals. Int J Cancer. 2007;121(4):759–766. doi: 10.1002/ijc.22769. [DOI] [PubMed] [Google Scholar]
- 34.Vazquez-Mena O, Medina-Martinez I, Juárez-Torres E, et al. Amplified genes may be overexpressed, unchanged, or downregulated in cervical cancer cell lines. PLoS One. 2012;7(3):e32667. doi: 10.1371/journal.pone.0032667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Espinosa AM, Alfaro A, Roman-Basaure E, et al. Mitosis is a source of potential markers for screening and survival and therapeutic targets in cervical cancer. PLoS One. 2013;8(2):e55975. doi: 10.1371/journal.pone.0055975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
The alternative splicing junctions of the four paired CSCC and ATN tissues
| Junction | Strand | Official gene name | LHR | LYM | ZTF | WSN |
|---|---|---|---|---|---|---|
| chr9:100767385–100773605 | + | ANP32B | (1/0) | (2/0) | (1/0) | (3/0) |
| chr1:111985966–111986338 | − | WDR77 | (1/0) | (2/0) | (1/0) | (2/0) |
| chr1:156015471–156018341 | − | UBQLN4 | (1/0) | (1/0) | (1/0) | (1/0) |
| chr1:156563314–156563723 | + | APOA1BP | (1/0) | (2/0) | (1/0) | (2/0) |
| chr1:160959034–160967990 | + | F11r | (1/0) | (8/0) | (2/0) | (3/0) |
| chr1:1770627–1784640 | − | GNB1 | (1/0) | (2/0) | (1/0) | (5/0) |
| chr1:241799325–241803233 | − | OPN3 | (2/0) | (1/0) | (1/0) | (6/0) |
| chr10:103769156–103771529 | − | c10orf76 | (2/0) | (1/0) | (2/0) | (1/0) |
| chr10:134143939–134145035 | − | LRRC27 | (1/0) | (1/0) | (2/0) | (2/0) |
| chr12:100574444–100589409 | − | microRNA 1827 | (1/0) | (1/0) | (2/0) | (1/0) |
| chr13:103436812–103440233 | − | Kdelc1 | (1/0) | (2/0) | (1/0) | (1/0) |
| chr13:77703315–77713380 | − | MYCBP2 | (1/0) | (2/0) | (1/0) | (4/0) |
| chr13:79889075–79893005 | − | RBM26 | (2/0) | (1/0) | (1/0) | (1/0) |
| chr14:103589770–103592696 | + | TNFAIP2 | (3/0) | (4/0) | (2/0) | (11/0) |
| chr15:30939867–30951018 | + | ARHGAP11B | (1/0) | (2/0) | (1/0) | (2/0) |
| chr15:68489916–68521889 | − | calml4 | (1/0) | (1/0) | (1/0) | (1/0) |
| chr15:93444004–93467600 | + | CHD2 | (2/0) | (2/0) | (1/0) | (2/0) |
| chr16:55463342–55478074 | − | MMP2 | (1/0) | (1/0) | (2/0) | (3/0) |
| chr19:13228895–13246063 | + | NACC1 | (1/0) | (1/0) | (1/0) | (1/0) |
| chr19:48653417–48657178 | − | lig1 | (1/3) | (2/0) | (1/0) | (1/0) |
| chr20:32224464–32228199 | + | CBFA2T2 | (2/0) | (2/0) | (1/0) | (1/0) |
| chr20:57484584–57485055 | + | Gnas | (1/0) | (1/0) | (1/0) | (1/0) |
| chr20:58450474–58453112 | − | sycp2 | (13/0) | (12/0) | (2/0) | (3/0) |
| chr22:24179991–24180800 | − | Derl3 | (2/0) | (1/0) | (1/0) | (4/0) |
| chr22:50981278–50983265 | − | KLHDC7B | (3/0) | (18/0) | (1/0) | (22/0) |
| chr4:119459113–119461423 | + | CEP170P1 | (0/2) | (0/3) | (0/2) | (0/5) |
| chr4:184255479–184255672 | − | CLDN24 | (1/0) | (2/0) | (1/0) | (1/0) |
| chr5:33466262–33467025 | + | Tars | (1/0) | (4/0) | (2/0) | (1/0) |
| chr6:52128956–52129242 | − | MCM3 | (4/0) | (3/0) | (2/0) | (3/0) |
| chr7:6066538–6068295 | − | Eif2ak1 | (1/0) | (1/0) | (1/0) | (1/0) |
| chr8:145666203–145667469 | − | TONSL | (1/0) | (1/0) | (1/0) | (1/0) |
| chr8:74872003–74884326 | − | TCEB1 | (2/0) | (1/0) | (2/0) | (1/1) |
| chr9:5436644–5438262 | − | PLGRKT | (1/0) | (1/0) | (2/0) | (1/0) |
| chr9:99086401–99122485 | − | SLC35D2 | (1/0) | (1/0) | (1/0) | (1/0) |
Abbreviations: ATN, adjacent nontumor tissues; CSCC, cervical squamous cell carcinoma.


