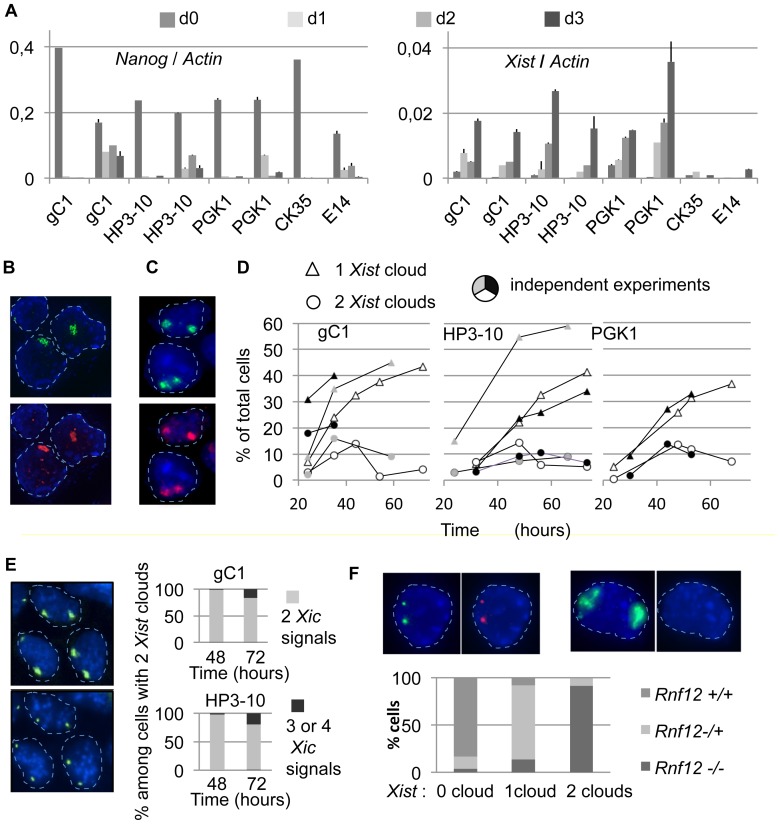
Figure 1. Initiation of X chromosome inactivation can occur on both Xs in the course of differentiation of naive ES cells.
A) Kinetic analysis using quantitative RT-PCR of the expression levels of Nanog and Xist normalized to Actin levels in the course of three days of differentiation of ES cells. Wild-type female cell lines: gC1, HP3-10 and PGK1. Control wild-type male cell lines: CK35 and E14Tg2A. RT- controls required at least 10 cycles more than the RT+ samples in order to exceed threshold. Columns represent the ratio of the means of triplicate measurement of 2-ct for Nanog, Xist and Actin at each timepoint in each differentiation experiment. Error bars represent standard deviation of the ratios of Xist (or Nanog) to Actin calculated as: sd of Xist/Actin = (mean of Xist/Actin) x [(sd of Xist/mean of Xist)2 + (sd of Actin/mean of Actin)2]0,5. Two fully independent differentiation experiments performed on separate days with each female ES cell line are shown. B) Immuno-RNA-FISH of Xist and H3-K27me3 using the HP3-10 cell line differentiated for 3 days: Xist RNA clouds (green, top panel) correspond to domains of enrichment for H3-K27me3 (red, bottom panel). In B, C, E, and F, DAPI blue staining was digitally dampened to favor visualization of the other channels. Nuclei are circled with a dashed line. C) Immuno-RNA-FISH of Xist and H3-K27me3 using the gC1 cell line differentiated for 36 hours. Nuclei showing two Xist clouds (green, top panel) additionally show two domains of enrichment for the H3-K27me3 mark (red, bottom panel). D) Kinetic analysis using Xist RNA-FISH. Cells showing one or two Xist clouds were counted over the course of independent 72-hours differentiation experiments with the gC1, HP3-10 and PGK1 ES cell lines (n>250). E) Differentiated female ES cells presenting two Xist clouds have a normal complement of two X chromosomes. Sequential RNA-FISH for Xist (top left panel) and DNA-FISH using a BAC-561P13 probe which maps within the X inactivation center (bottom left panel) were performed with the HP3-10 cell line. This experiment was similarly performed with the gC1 ES cell line. Cells presenting two Xist clouds were evaluated for their complement of X chromosomes (right panels; gC1 48 h, n = 60; gC1 72 h, n = 25; HP3-10 48 h; n = 45; HP3-10 72 h, n = 30). All or most cells presenting two Xist clouds have a normal complement of two X chromosomes, although some X triplody arouse at 72 hours in this cell population. F) Double RNA-FISH for Xist (green) and Rnf12 (red) using the HP3-10 ES cell line differentiated during 40 hours. A cell which did not upregulate Xist expressed Rnf12 bi-allelically (left panels) and a cell which upregulated both alleles of Xist silenced Rnf12 (right panels). The efficiency of detection of the Rnf12 primary transcription site was 86% as determined in cells lacking Xist expression. Rnf12 expression was examined in cells with no Xist cloud (n = 159), with one Xist cloud (n = 181) or with 2 Xist clouds (n = 75). Nearly all the cells presenting 2 Xist clouds were functionally nullisomic for Rnf12 expression. Repeat experiments with the HP3-10 and gC1 ES cell lines gave essentially identical results.