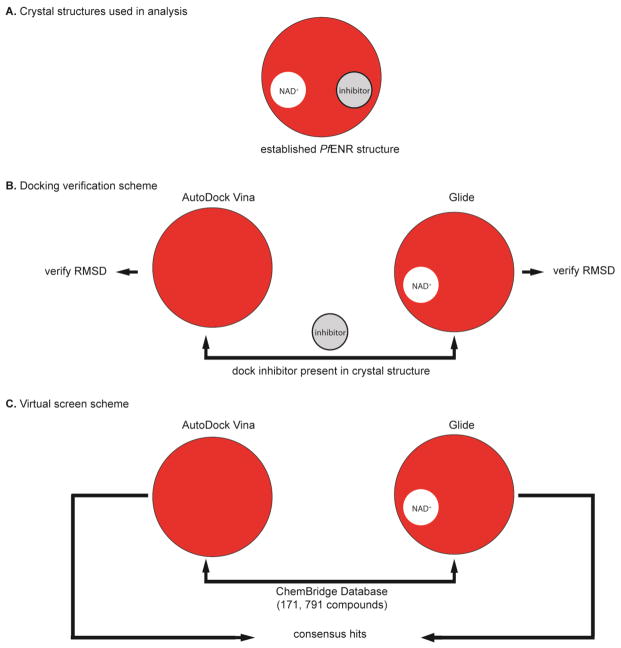
Figure 3.
Virtual screen workflow for identifying in silico small molecule hits for in vitro assay. (A) Three established PfENR structures were chosen for docking (PDB: 1UH5, 3LSY, 3LT0). (B) The docking protocols were evaluated for their ability to dock known inhibitors of PfENR. Inhibitors and cofactors were removed from the AutoDock Vina receptor files, while only inhibitors were removed from the Glide receptor grid files. The inhibitors were re-docked into these prepared structures and evaluated for their ability to dock known inhibitors by calculating the RMSD. (C) After establishing a validated docking program, 171,791 compounds from the ChemBridge EXPRESS-Pick compound collection were docked into the prepared ENR structures and a consensus in silico small molecule hit set was generated for in vitro testing.