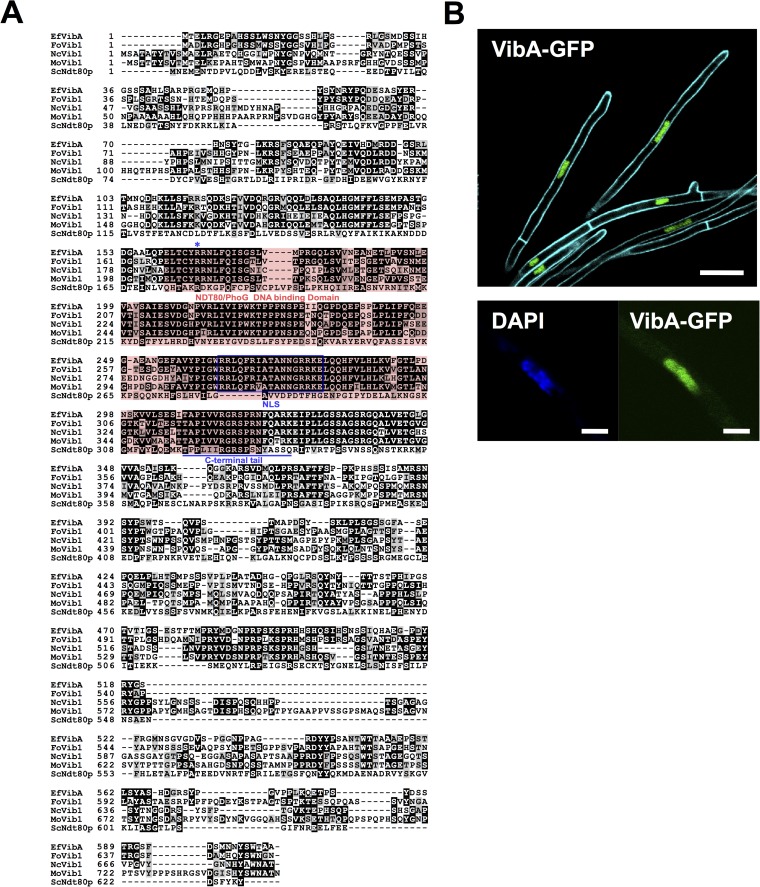
FIG 3.
(A) The deduced protein sequence of E. festucae VibA (EfVibA) was aligned with those of Fusarium oxysporum Vib1 (FoVib1; accession no. FOXB_17342), Magnaporthe oryzae Vib1 (MoVib1; accession no. MGG_00729), Neurospora crassa Vib1 (NcVib1; accession no. NCU03725.5), and Saccharomyces cerevisiae Ndt80p by ClustalW ver. 2.1 (34) with default settings. The NDT80/PhoG DNA binding domain is indicated with a red box. A predicted bipartite nuclear localization signal (NLS) is indicated in a blue box. The C-terminal tail of Ndt80p predicted to make contact with the DNA major groove (54) is underlined, and the arginine residue required for the function of Ndt80p in Candida albicans (55) is indicated by an asterisk. (B) Localization of VibA-GFP in hyphae of E. festucae. VibA-GFP was expressed under the control of a TEF promoter in E. festucae E437 and then monitored by confocal laser scanning microscopy. (Bottom) Hypha of E. festucae expressing VibA-GFP and stained with DAPI. Bars = 10 μm (upper panel) and 2 μm (lower panel).