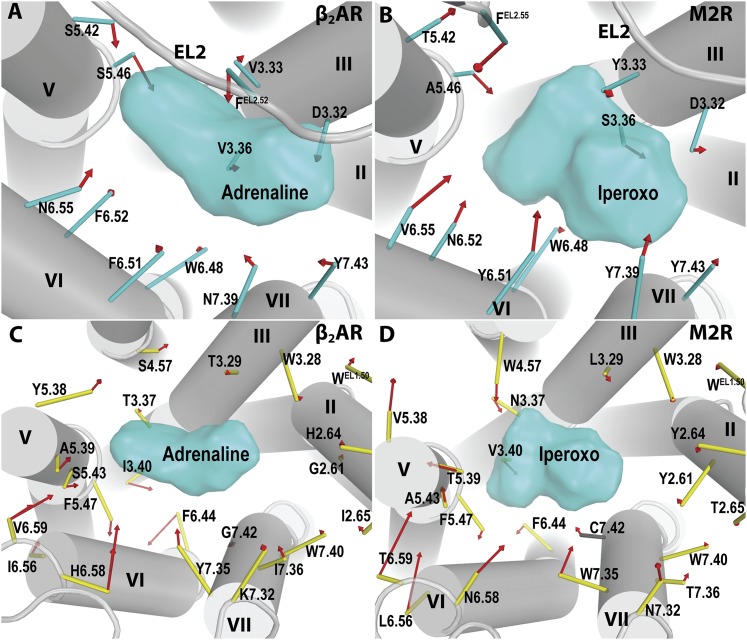
Fig. 8.
Rearrangements of binding site residues in the conformational transition from the inactive state to the active state. The rearrangements in the OBS (A and B) and SBPs (C and D) between the inactive and active structures of β2AR (PDB IDs 2RH1 and 4LDE/4LDO) (A and C) and M2R (PDB IDs 3UON and 4MQT) (B and D) are shown. The color coding of residues is the same as in Fig. 1. For simplicity, EL2 is not shown in (C and D). The agonists bound in the active-state structures are represented as cyan surfaces to indicate the space occupied by the OBS. For clarity, only the backbones of the inactive-state structures are shown in gray cartoon representation, whereas the active-state structures are superimposed to the corresponding inactive-state structures by the Cα atoms of 70 TM residue positions that undergo the smallest changes between the inactive and active states. To select these positions, we ranked the distance changes of the corresponding Cα atoms in the inactive and active structures, after aligning them by TM residues using the iterative-fit “align” command in PyMOL (version 1.3r1; Schrödinger LLC, New York, NY). This ranking has been carried out for both β2AR and M2R structures pairs; the 70 highest ranked positions were selected after averaging the ranks from both receptor comparisons and ensuring that at least two positions from each TM were included. The ligand-binding site residues are represented as sticks, drawn from the Cα atom to the COM of the side chain heavy atoms of the residue (for Gly, only the Cα atom is shown). The red arrows indicate the conformational changes from the inactive to the active state for the COM of each residue. In (C), two arrows are drawn for each of the positions 6.58 and 6.59, showing the differing conformational changes to the BI-167107–bound active-state (PDB ID 4LDE) compared with the adrenaline-bound active-state (PDB ID 4LDO) for β2AR.