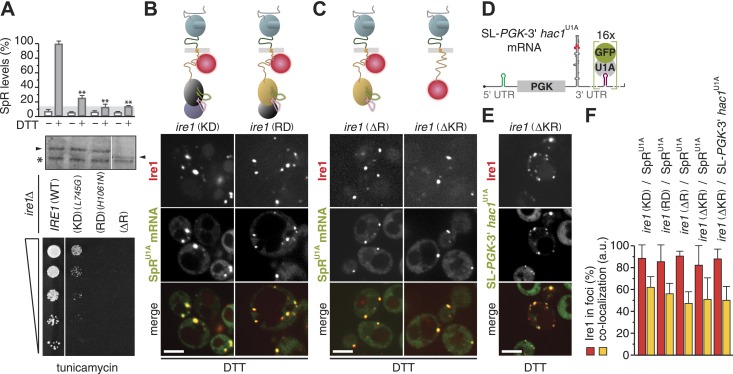
Figure 2. The kinase and RNase domains of Ire1 are dispensable for foci formation and mRNA recruitment.
(A) Splicing reporter assay before or after ER-stress induction with 2 mM DTT for 2 hr (top), Western blot of Ire1 (middle), and viability assay under ER-stress conditions (0.2 µg ml−1 tunicamycin; bottom) were performed in ire1Δ yeast containing a genomic copy of the SpR, complemented with wild-type (WT), kinase dead (KD), RNase dead (RD), and RNase truncation (ΔR) mutant alleles of ire1. Maximal (100%) and background level (14%, light gray bar) fluorescence are set as in Figure 1A. Mean and s.d. are shown (n = 2). Statistical significance in a Student's t-test of differences in splicing levels as compared with wild-type is indicated (**p ≤ 0.01). The arrowheads denote (mutant or truncated) Ire1 protein and the asterisk a background band on the immunoblot as in Figure 1H. (B, C) Top: schematic of the mCherry-tagged versions of the same ire1 mutants as in (A) as well as a kinase/RNase truncation (ΔKR) mutant, color-coded as in Figure 1A, except defective domains are black. (D) Schematic of a chimeric mRNA, SL-PGK1-3′ hac1U1A, which is PGK1U1A, bearing in its 3′ UTR the stem-loop structure with the 3′ BE of the HAC1 mRNA and in its 5′ UTR a small stem-loop (green) that confers translational repression (Aragón et al., 2009). (B, C, E) Localization of Ire1–mCherry and of U1A–GFP decorating either SpRU1A (B, C) or SL-PGK1-3′ hac1U1A (E) mRNA. ER-stress was induced with 10 mM DTT for 45 min; imaging was performed of ire1Δ cells, complemented with Ire1 imaging constructs, as depicted. Scale bars represent 5 µm. (F) Bar diagrams depict the percentage of Ire1 signal in foci (red bars) and the co-localization index for mRNA recruitment into foci of Ire1 variants shown in B, C, and E (mean and s.e.m., n = 5–10). There is no statistical significance in a Student's t-test of differences in foci formation and mRNA recruitments as compared with wild-type.
DOI: http://dx.doi.org/10.7554/eLife.05031.005