Abstract
Altered microRNA (miRNA) associated with gastric cancer (GC) development and miR-17 and miR-106b were differentially expressed in GC tissues. This study detected serum levels of miR-17 and miR-106b expression in GC, benign gastric disease (BGD) and healthy controls to assess them as tumor markers for GC. Serum samples from 40 GC, 32 BGD (10 gastric ulcer, 14 gastric polyps, and 8 gastric ulcer with polyps) and 36 healthy individuals were subjected to quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of miR-17 and miR-106b expression. The data showed that the serum levels of miR-17 and miR-106b were significantly reduced in healthy individuals and BGD patients compared to GC patients. There was a significant association of miR-17 and miR-106b expression with age, but not with other clinicopathological features, such as gender, tumor differentiation, stage and lymphatic metastasis. Further analysis showed that, in discriminating GC patients from healthy controls, miR-17 could yield a receiver-operating characteristic (ROC) area under the curve (AUC) of 0.879 with 80.6% sensitivity and 87.5% specificity and miR-106b could yield an AUC of 0.856 with 75.0% sensitivity and 92.5% specificity. The combined AUC of miR-17 and miR-106b was 0.913 with 83.3% sensitivity and 87.5% specificity. Collectively, these data suggest that detection of serum miR-17 and miR-106b levels should be further evaluated as novel non-invasive biomarkers in early GC detection and surveillance of disease progression.
Keywords: miR-17, miR-106b, gastric cancer (GC), benign gastric disease (BGD), biomarker
Introduction
MicroRNA (miRNA) is a class of naturally occurring small noncoding RNAs that are 18 to 22 nucleotides in length and function to post-transcriptionally silence protein expression by binding to complementary target messenger RNAs, thereby degrading these messenger RNAs or inhibiting them from being translated into proteins. Aberrant miRNA expression promotes oncogenesis and the progression of various human cancers (1). In 2008, Mitchell et al. (2) first evaluated blood-circulating miRNAs and demonstrated that they could be used as non-invasive biomarkers to detect prostate cancer. A great number of studies have since revealed that various blood-circulating miRNAs could be candidate biomarkers in different cancers, including gastric cancer (GC) (3,4). Other studies showed that changes in the serum levels of miRNA expression were the opposite of those observed in tissue samples (3,5,6). However, the underlying mechanism and cause of these changes remain to be determined.
A number of previous studies have shown that miR-17 and miR-106b expression was upregulated in GC tissue specimens and detection of their expression could distinguish GC tissues from adjacent noncancerous tissues (7-9). However, to date, few studies have reported serum levels of miR-17 and miR-106b in GC in the literature. Furthermore, no study has compared serum levels of miR-17 and miR-106b among normal, benign gastric disease (BGD) and GC patients.
As we know, GC development is a dynamic process that progresses from normal mucosae through BGD to gastric carcinogenesis. Thus, this study assessed serum levels of miR-17 and miR-106b in GC, BGD and healthy controls to evaluate them as tumor markers for the early diagnosis of GC.
Materials and methods
Patients and serum samples
A total of 108 subjects, including 40 GC patients, 32 BGD patients (10 gastric ulcer patients, 14 gastric polyp patients, and 8 patients with gastric ulcer and polyps) and 36 healthy controls were recruited to this study. BGD patients and healthy controls matched with the age and sex of GC patients were recruited. Whole blood samples were collected from patients before surgery, radiotherapy and chemotherapy, as well as from healthy volunteers at the Third Xiangya Hospital, the Central South University between October 2012 and August 2014. The Ethnic Committee of the hospital approved the study, and the informed consent of all the participants was obtained.
RNA isolation, cDNA synthesis and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Blood samples were left at room temperature for 1 h after collection for coagulation and then centrifuged at 2,000 r/min for 10 min, 3,000 r/min for 5 min and 4,500 r/min for 5 min at 4 °C. Serum samples were then collected and stored at –80 °C until use.
To detect the serum levels of miRNA expression, we isolated total RNA from 250 µL serum using a total RNA extraction kit (QuantoBio, Beijing, China) according to the manufacture’s protocol. The concentration of RNA was measured by a spectrophotometer and ranged from 4.7 to 25.8 ng/µL per serum sample. After that, miRNAs were polyadenylated by poly(A) polymerase and reverse transcribed to cDNA using a Reverse Transcriptase Kit (QuantoBio) following the manufacture’s protocol.
qPCR amplification was performed in the Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems, Frost city, CA, USA) using the qPCR detection kit from QuantoBio. QuantoBio also supplied the miRNA-specific primers. The reaction was set up using a 20 µL volume protocol: 1 µL cDNA, 2 µL specific primer, 10 µL 2× qRCR Mix, 2 µL 10× UPM, 0.4 µL ROX and 4.6 µL Milli-Q-water. The reaction mixtures were incubated for an initial 95 °C for 5 min, followed by 40 cycles of denaturation for 15 s at 95 °C, annealing for 15 s at 55 °C and extension for 30 s at 60 °C. At the end of the PCR cycles, the melting curve analysis was performed to validate the specificity of the expected PCR product. Each reaction was carried out in triplicate and QuantoEC-2 (QuantoBio) was used as a normalization control. Serum level of miRNA was calculated using the 2–∆∆Ct method relative to QuantoEC-2.
Statistical analysis
The data were analyzed for analysis of variance, Student-Newman-Keuls test, Kruskal-Wallis test and Mann-Whitney U test using SPSS 19.0 software (SPSS Inc., Chicago, IL, USA). P<0.05 was considered statistically significant. Receiver-operating characteristic (ROC) curves and area under the curve (AUC) were calculated to assess the feasibility of miRNA as a diagnostic biomarker for sensitivity and specificity using SPSS 19.0 software in a non-parametric way. The ROC curve, a graphical plot, can illustrate the performance of a binary classifier system at various threshold settings to calculate the fraction of true positivity out of the total actual positive rate vs. the fraction of false positivity for a selected sensitivity and specificity for a given biomarker.
Results
Reduced serum levels of miR-17 in GC patients compared to BGD patients and healthy controls
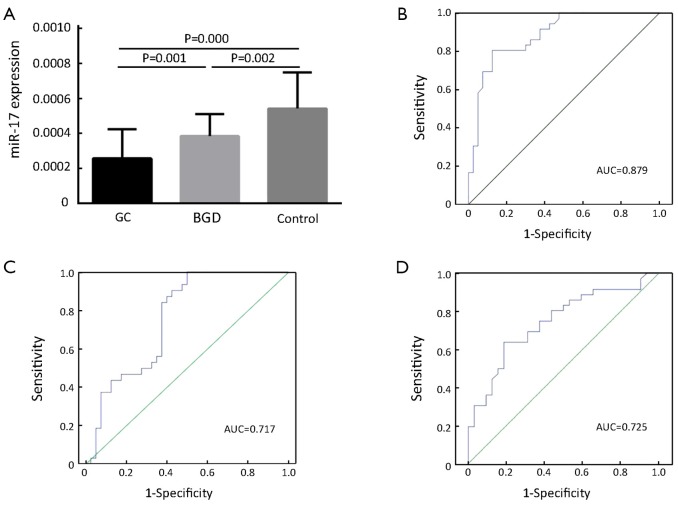
As shown in Figure 1A, the serum levels of miR-17 were significantly reduced in both GC and BGD patients compared to those of healthy controls (P=0.000 and 0.002, respectively) and significantly lower in GC patients than in BGD patients (P=0.001). Moreover, we also performed ROC curve analysis to evaluate the diagnostic value of miR-17 and found an AUC of 0.879 with 80.6% sensitivity and 87.5% specificity for discriminating GC patients from controls (Figure 1B), an AUC of 0.717 with 90.6% sensitivity and 57.5% specificity for discriminating GC patients from BGD patients (Figure 1C) and an AUC of 0.725 with 62.9% sensitivity and 81.2% specificity for discriminating BGD patients from controls (Figure 1D).
Figure 1.
Reduced levels of serum miR-17 in GC compared to healthy control and BGD. qRT-PCR was performed to detect serum levels of miR-17 in 40 GC, 32 BGD and 36 healthy control samples. (A) The concentration of miR-17 was significantly lower in sera from both GC and BGD patients than in that from healthy individuals (P=0.000 and P=0.002, respectively). Serum levels of miR-17 were significantly lower in GC patients than in BGD patients (P=0.001). Serum levels in order of expression level were healthy control >BGD >GC; (B) ROC curve analysis reveals an AUC of 0.879 in discriminating GC patients from controls; (C) ROC curve analysis reveals an AUC of 0.717 in discriminating GC patients from BGD patients; (D) ROC curve analysis reveals an AUC of 0.725 in discriminating BGD patients from controls. GC, gastric cancer; BGD, benign gastric disease; qRT-PCR, quantitative reverse transcription polymerase chain reaction; ROC, receiver-operating characteristic; AUC, area under the curve.
Reduced serum levels of miR-106b in GC patients compared to BGD patients and healthy controls
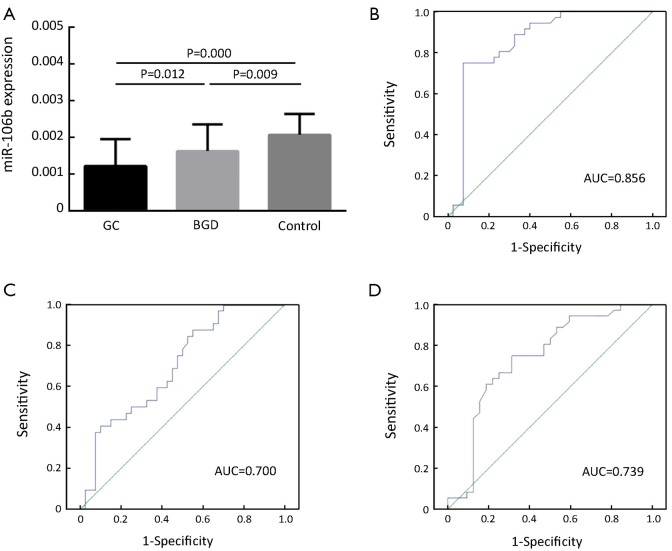
The serum levels of miR-106b were also significantly reduced in both GC and BGD patients compared to those of healthy controls (P=0.000 and 0.009, respectively, Figure 2A) and were significantly lower in GC patients than in BGD patients (P=0.012, Figure 2A). Moreover, the ROC curve analysis revealed an AUC of 0.856 with 75.0% sensitivity and 92.5% specificity for discriminating GC patients from controls (Figure 2B), an AUC of 0.700 with 87.2% sensitivity and 45.0% specificity for discriminating GC from BGD patients (Figure 2C) and an AUC of 0.739 with 75.0% sensitivity and 68.7% specificity for discriminating BGD patients from controls (Figure 2D).
Figure 2.
Reduced levels of serum miR-106b in GC compared to healthy control and BGD. qRT-PCR was performed to detect serum levels of miR-106b in 40 GC, 32 BGD and 36 healthy control samples. (A) The concentration of miR-17 was significantly lower in sera from both GC and BGD patients than in that from healthy individuals (P=0.000 and P=0.009, respectively). Serum levels of miR-17 were significantly lower in GC patients than BGD patients (P=0.012). Serum levels in order of expression level were healthy control>BGD>GC; (B) ROC curve analysis reveals an AUC of 0.856 in discriminating GC patients from controls; (C) ROC curve analysis reveals an AUC of 0.700 in discriminating GC patients from BGD patients; (D) ROC curve analysis reveals an AUC of 0.739 in discriminating BGD patients from controls. GC, gastric cancer; BGD, benign gastric disease; qRT-PCR, quantitative reverse transcription polymerase chain reaction; ROC, receiver-operating characteristic; AUC, area under the curve.
Furthermore, we used combined miR-17 and miR-106b serum levels to improve the sensitivity and specificity of GC detection. The data showed that the AUC of combined miR-17 and miR-106b was 0.913 with 83.3% sensitivity and 87.5% specificity for discriminating GC patients from controls (Figure 3).
Figure 3.
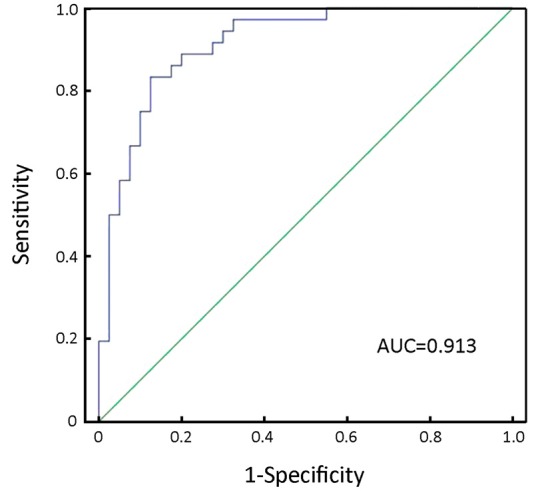
ROC curve analysis of combined miR-17 and miR-106b serum levels in GC detection. The combined serum levels of miR-17 and miR-106b yield an AUC of 0.913 with 83.3% sensitivity and 87.5% specificity in discriminating GC patients from controls. ROC, receiver-operating characteristic; GC, gastric cancer; AUC, area under the curve.
Association of serum miR-17 and miR-106b levels with clinicopathological features of GC patients
To discern whether the serum levels of miR-17 and miR-106b were connected with the clinicopathological features of GC patients, we separated these patients by gender, age, tumor differentiation, stage and lymphatic metastasis. As shown in Table 1, the expression of miR-17 and miR-106b was associated with patient age (P<0.05), but not associated with gender, tumor differentiation, TNM stage or lymph node metastasis (P>0.05 using Kruskal-Wallis test). The expression of both miR-17 and miR-106b was significantly lower in patients with older ages.
Table 1. Association of serum miR-17 and miR-106b levels with clinicopathological features of GC patients.
| Clinicopathological characteristics | N | P |
|
|---|---|---|---|
| miR-17 | miR-106b | ||
| Gender | 0.666 | 0.896 | |
| Male | 28 | ||
| Female | 12 | ||
| Age (years) | 0.02 | 0.017 | |
| >53 | 26 | ||
| ≤53 | 14 | ||
| Tumor differentiation | 0.857 | 0.847 | |
| Well | 10 | ||
| Poor | 30 | ||
| TNM stage | 0.602 | 0.402 | |
| Early (I/II) | 9 | ||
| Later (III/IV) | 31 | ||
| Lymph node metastasis | 0.97 | 0.83 | |
| Yes | 28 | ||
| No | 12 | ||
GC, gastric cancer.
Conclusions
Our current findings suggest that the serum levels of miR-17 and miR-106b are highest in healthy individuals, significantly decline in BGD patients and are expressed at an even lower level in GC patients. Thus, miR-17 and miR-106b are promising non-invasive biomarkers for the early diagnosis of GC and disease progression surveillance, and they should be further evaluated for clinical use. The serum levels of both miR-17 and miR-106b have moderate sensitivity and specificity in GC diagnosis; however, since the level of serum miR-17 and miR-106b expression was reduced in GC, combination with other biomarkers, like carcinoembryonic antigen (CEA), could yield improved sensitivity and specificity in GC diagnosis. Moreover, older GC patients had lower levels of serum miR-17 and miR-106b expression, indicating that miR-17 and miR-106b might be important regulators on the crossroads between aging and GC. Future studies will focus on developing a clinically useful early diagnostic test for GC using these biomarkers and will validate our current data in a larger sample size study.
Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No. 81372140, 81301688, 81272192, 81171882, 81172298); Ph.D. Programs Foundation of Ministry of Education of China (No. 20130162110050 and 20130162120093); Post-doctoral Foundation of Central South University (No. 131425); China Postdoctoral Science Foundation (2014M552167); Key Program for International Cooperation Projects of Hunan Province (no. 2011WK2011); Natural Science Foundation of Hunan Province (Grant No. 12JJ4088); Technology Project of Hunan Province (2012SK3229); Research foundation of Health Department of Hunan Province (B2012-100); 125 Talent Project of the Third Xiangya Hospital of Central South University.
Disclosure: The authors declare no conflict of interest.
References
- 1.Su X, Xing J, Wang Z, et al. microRNAs and ceRNAs: RNA networks in pathogenesis of cancer. Chin J Cancer Res 2013;25:235-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu H, Zhu L, Liu B, et al. Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer Lett 2012;316:196-203. [DOI] [PubMed] [Google Scholar]
- 4.Ma YY, Tao HQ. Microribonucleic acids and gastric cancer. Cancer Sci 2012;103:620-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka M, Oikawa K, Takanashi M, et al. Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PLoS One 2009;4:e5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang K, Zhang S, Marzolf B, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A 2009;106:4402-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang JL, Hu Y, Kong X, et al. Candidate microRNA biomarkers in human gastric cancer: a systematic review and validation study. PLoS One 2013;8:e73683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park D, Lee SC, Park JW, et al. Overexpression of miR-17 in gastric cancer is correlated with proliferation-associated oncogene amplification. Pathol Int 2014;64:309-14. [DOI] [PubMed] [Google Scholar]
- 9.Yang TS, Yang XH, Chen X, et al. MicroRNA-106b in cancer-associated fibroblasts from gastric cancer promotes cell migration and invasion by targeting PTEN. FEBS Lett 2014;588:2162-9. [DOI] [PubMed] [Google Scholar]