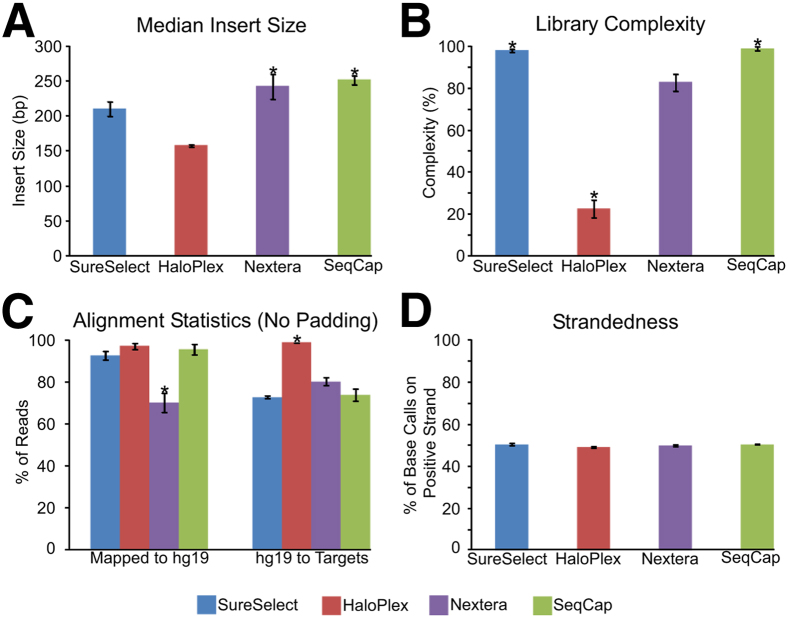
Figure 2.
Comparison of various library metrics. A: Library insert size (median ± median absolute deviation throughout figure). B: Library complexity, as measured by percentage of unique reads (of the total reads) in each library. C: Alignment metrics for each technology for reads (ie, after removing duplicates for SureSelect, Nextera, and SeqCap) mapping to hg19 and to each technology's targeted regions. D: Strandedness for each technology, as measured by the number of base calls on the positive strand. ∗P = 3.82 × 10−4 (U-test), Nextera and SeqCap have a significantly higher insert size than SureSelect and HaloPlex (A); ∗P < 9.24 × 10−6, SureSelect and SeqCap have significantly greater complexity than HaloPlex and Nextera, whereas HaloPlex has the lowest complexity (B); ∗P < 10−5, Nextera has the lowest alignment rate to the genome and HaloPlex has the highest alignment rate to target regions (C).