Summary
Viral pathogenesis relies upon modulation of host cytokine activation as well as cell death pathways. Infection by murine cytomegalovirus induces a novel receptor-interacting protein (RIP)3-dependent necrosis. RIP3 kinase activity and homotypic interaction motif (RHIM)-dependent interactions control virus-associated necrosis as occurs in TNFα-induced necroptosis; however, the virus-induced death pathway proceeds independent of RIP1, and is therefore distinct from the TNFα-dependent death pathway. The viral inhibitor of RIP activation (vIRA, encoded by the viral M45 gene) suppresses either pathway, disrupting RHIM-dependent RIP3-RIP1 interaction that is critical for TNFα-induced necroptosis as well as a RIP3 RHIM-dependent step in virus-induced necrosis. Importantly, the attenuation of vIRA-deficient virus in wild type mice is completely normalized in a RIP3-deficient genetic background. Thus, vIRA function validates necrosis as central to the elimination of infected cells in host defense and highlights the benefit of multiple virus-encoded cell death suppressors that subvert not only apoptotic, but also necrotic mechanisms of virus clearance.
Keywords: herpesvirus, cytomegalovirus, cell death suppressor, necroptosis, pathogen sensor
Introduction
Cellular sensing of viral pathogens by the host activates inflammatory gene expression and triggers cell death. These distinct cell-intrinsic response pathways directly control viral spread from the portal of entry and influence the quality of pathogen-specific adaptive immunity. Innate antiviral cytokines such as interferon (IFN) have long been considered central to control of viral spread (McCartney and Colonna, 2009; Roy and Mocarski, 2007; Takeuchi and Akira, 2009), with caspase-dependent apoptotic cell death viewed as a complementary, evolutionarily conserved clearance mechanism triggered by distinct signals. The importance of death has been reinforced by the widespread existence of apoptotic cell death suppressors, including those that viruses employ to subvert intrinsic clearance (Clem, 2005; D’Agostino et al., 2005; Kepp et al., 2009). These include (i) caspase inhibitors, such as baculovirus p35 (Bump et al., 1995; Clem et al., 1991), viral inhibitors of apoptosis (IAPs) (Crook et al., 1993), poxvirus crmA (Miura et al., 1993; Ray et al., 1992) and viral FLICE (caspase-8) inhibitory proteins (FLIPs) (Thome et al., 1997), as well as, (ii) mitochondrial cell death suppressors such as viral Bcl-2 homologs and other proteins encoded by large DNA viruses that block cytochrome c release from mitochondria (Galluzzi et al., 2008; Goldmacher, 2005; White, 2006). Although apoptosis is a well-established cell-intrinsic response to pathogens, caspase-independent cell death, or programmed necrosis, has recently emerged as an alternative death pathway that dominates under specific conditions (Festjens et al., 2007; Hitomi et al., 2008). Necroptosis is a form of programmed necrosis induced by death receptors (DR), a subgroup of the tumor necrosis factor (TNF) superfamily, that is independent of caspases but dependent on the activity of the adaptors receptor interacting protein kinase 1 (RIP1) (Festjens et al., 2007; Meylan and Tschopp, 2005) and RIP3 (Cho et al., 2009; He et al., 2009; Zhang et al., 2009), two related RIP homotypic interaction motif (RHIM)-containing protein kinases (Sun et al., 2002). Programmed necrosis has been viewed as a back-up to apoptosis (Festjens et al., 2007; Festjens et al., 2006) because this pathway is activated by the same signals under experimental conditions where caspase-8 activation has been suppressed or is absent. It has recently been proposed that this alternative death pathway is another host counter-measure against invading pathogens (Cho et al., 2009). However, suppression of this pathway by a microbe for pathogenesis in vivo has yet to be established.
RIP1, the founding member of a serine-threonine protein kinase family related to interleukin 1 receptor-associated kinases, transduces inflammatory and cell death signals in cells following DR ligation, activation of pattern recognition receptors (PRRs), and DNA damage (Janssens et al., 2005; Meylan et al., 2004; Michallet et al., 2008; Stanger et al., 1995). An amino-terminal kinase domain in RIP1 is critical for DR-induced necroptosis (Holler et al., 2000) and is the target of necrostatin-1 (Nec-1) (Degterev et al., 2008; Degterev et al., 2005), a defining inhibitor of necroptosis. Other RIP family members share a kinase domain but carry different protein-protein interaction motifs. RIP1 has a carboxyl-terminal death domain (DD) that engages other DD-containing proteins and transduces signals from death receptors (Stanger et al., 1995) as well as a central (intermediate) domain important for NF-κB activation (Festjens et al., 2007) and RHIM-dependent signaling (Sun et al., 2002). Three other cellular RHIM-containing adaptors interact with RIP1 to initiate gene activation and death pathways: (i) RIP3, the only other RHIM-containing RIP family member (Sun et al., 2002), (ii) the toll-like receptor (TLR)3 and TLR4 adaptor TIR domain-containing adaptor-inducing IFNβ (TRIF) (Kaiser and Offermann, 2005; Meylan et al., 2004), and (iii) DNA-dependent activator of IFN-regulatory factors (DAI, also termed DLM-1 or ZBP1) (Kaiser et al., 2008; Rebsamen et al., 2009), a candidate DNA sensor (Takaoka et al., 2007). RIP1 is recruited to TLR3 and TLR4 by TRIF (Meylan et al., 2004) via RHIM-dependent interactions. Signaling via TLR3, TLR4 or DAI activates proinflammatory, NF-κB- and IRF3-dependent responses to endosomal dsRNA, lipopolysaccharide (LPS) or cytoplasmic dsDNA, respectively (Alexopoulou et al., 2001; Kawai and Akira, 2007; Takaoka et al., 2007). Thus, RIP1 is an important adaptor, balancing cellular signaling that results in inflammatory cytokine activation and influencing the initiation of programmed cell death pathways (Degterev and Yuan, 2008; Festjens et al., 2007; Meylan and Tschopp, 2005).
When first evaluated, RIP3 appeared to exert a negative modulatory role in TLR3-dependent or TNFα receptor NF-κB activation (Meylan et al., 2004; Sun et al., 2002). More recently, RIP3 has been ascribed a positive impact on DAI-induced, RIP1 RHIM-dependent NF-κB activation (Kaiser et al., 2008; Rebsamen et al., 2009) as well as in the RHIM-dependent initiation of necroptosis induced by TNFα (Cho et al., 2009; He et al., 2009; Zhang et al., 2009). RIP3- and RIP1-dependent necroptosis is triggered by death receptor stimulation when caspases are inhibited or absent (Ch’en et al., 2008; Cho et al., 2009; He et al., 2009; Holler et al., 2000; Vercammen et al., 1998; Yu et al., 2004; Zhang et al., 2009), although the requirement for caspase inhibition to unveil necroptosis has limited observations of this cell death pathway in natural biological settings (Festjens et al., 2006). RIP3-deficient mice are viable and display no obvious developmental phenotypes or defects in NF-κB signaling triggered by LPS or TNFα (Newton et al., 2004). One recent report highlighted decreased inflammation and liver pathology in vaccinia virus-infected RIP3-deficient mice, processes that were ascribed to altered TNFα-induced necroptosis (Cho et al., 2009).
As obligate intracellular pathogens that remain latent for life, herpesviruses are heavily vested in cell fate decisions. Human cytomegalovirus (HCMV), the prototypic, medically significant β-herpesvirus, and murine cytomegalovirus (MCMV), a surrogate used to model viral pathogenesis, encode several cell death suppressors (Cicin-Sain et al., 2008; Goldmacher et al., 1999; Mack et al., 2008; McCormick et al., 2005; McCormick et al., 2008; Menard et al., 2003; Skaletskaya et al., 2001; Upton et al., 2008). The MCMV M45-encoded viral inhibitor of RIP activation (vIRA) is a viral structural protein (Lembo et al., 2004; Kattenhorn et al., 2004) originally identified as a tropism determinant required for endothelial cell-specific viral replication (Brune et al., 2001). MCMV mutants lacking vIRA induce premature death in some cell lines, although the mode and mechanism of death remains controversial. Both caspase-dependent and caspase-independent cell death pathways have been implicated (Brune et al., 2001; Mack et al., 2008), leaving the question of mechanism open for resolution. Recent reports showing the critical role of RIP1 and RIP3 in DR-induced necroptosis (Cho et al., 2009; He et al., 2009; Zhang et al., 2009), together with previous evidence that vIRA physically interacts efficiently with RIP3 as well as RIP1 (Upton et al., 2008), implicate vIRA suppression of cell death pathways via modulation of RIP3 and/or RIP1 to provide cell death suppression in sensitive cell types and enhance virus replication and dissemination. However, the specific contributions of RIP1-RIP3 interactions to the mutant virus phenotype have not been established. Here, we demonstrate that vIRA disrupts both the RHIM-dependent RIP3-RIP1 kinase complex that initiates TNFα-induced necroptosis as well as a novel RIP3 RHIM-dependent, RIP1-independent step in virus-induced necrosis. Importantly, the severe attenuation of vIRA-deficient virus is normalized when mice lack RIP3 function. These data demonstrate that RIP3 mediates a necrotic death pathway independent of RIP1 and implicates viral control of this pro-necrotic adaptor protein in viral pathogenesis.
Results
vIRA RHIM-dependent interactions suppress virus induced programmed necrosis and are required for pathogenesis
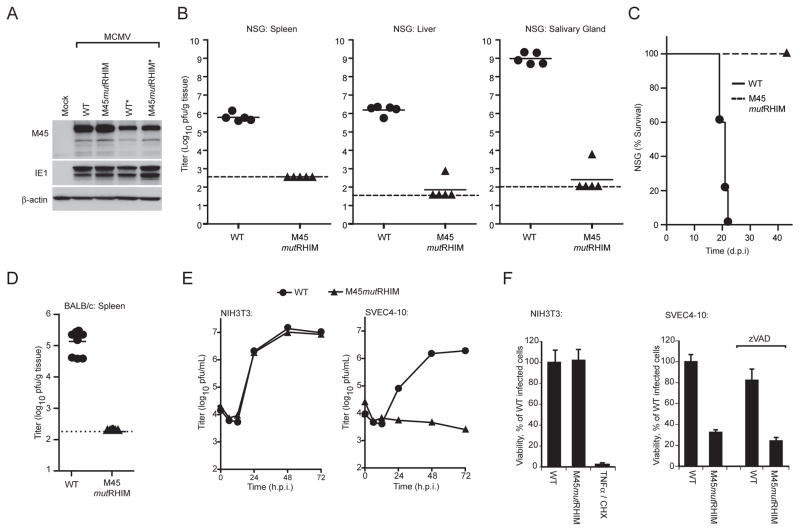
In order to explore the biological consequences of vIRA RHIM-specific interactions during viral infection, we generated a recombinant bacmid-derived mutant MCMV and evaluated its behavior in infected cells and animals. M45mutRHIM virus carried a tetra-alanine substitution (Figure S1A, B) previously shown to eliminate the capacity for this protein to engage in RHIM-dependent interactions (Upton et al., 2008). Viral stocks were recovered by transfection of bacmid DNA into NIH3T3 fibroblasts, where mutant and wild type (WT) exhibited equivalent replication properties as elaborated previously (Brune et al., 2001; Mack et al., 2008). Viral genome integrity was affirmed by restriction enzyme digestion of bacmid and viral DNA (Figure S1C, data not shown). Immunoblot analysis indicated MCMV mutant M45mutRHIM and WT viruses produced M45-proteins of comparable size and at similar levels of expression (Figure 1A). All samples contained equivalent amounts of β-actin as a loading control, and infected cells all expressed similar levels of viral immediate early (IE)1 protein. To investigate whether the in vivo behavior of vIRA RHIM-dependent interactions recapitulated the severe attenuation previously demonstrated for vIRA using null mutants (Lembo et al., 2004), we evaluated mutant and WT virus replication in immunodeficient as well as in immunocompetent mice. To evaluate the properties of viral replication, dissemination, and virulence independent of innate natural killer (NK) as well as adaptive T and B lympocyte function, the behavior of WT and M45mutRHIM viruses was compared in severely immunocompromised nonobese diabetic (NOD), severe combined immunodeficient (scid), IL2 common γ chain−/− (NSG) mice (Shultz et al., 2005). Mutant virus exhibited at least 1,000-fold reduced levels in spleen, 10,000-fold reduced levels in liver and 1,000,000-fold reduced levels in salivary glands compared to WT virus (Figure 1B). To investigate viral pathogenesis, cohorts of NSG mice inoculated with WT or mutant virus were followed for 6 weeks. While mice infected with WT virus succumbed between 20 to 22 days post-infection, mutant virus infected animals showed no signs of CMV-disease up to 42 days (Figure 1C) and beyond (data not shown). We extended in vivo studies to include immunocompetent mice, demonstrating that WT was detected in the spleens of BALB/c mice at day 3 postinoculation when virus titers normally peak (Figure 1D); whereas mutant virus remained undetectable at day 3 (Figure 1D) as well as at day 5 (data not shown). These data show that the phenotype of the M45mutRHIM mutant virus is similar to previously characterized vIRA-deficient viruses (Lembo et al., 2004). Importantly, the attenuated phenotype of the tetra-alanine substitution mutant in vivo established the critical role of RHIM-dependent interactions in vIRA function, extending prior investigations that relied upon viral mutants which truncated or eliminated vIRA expression. This result implicates vIRA RHIM-dependent interaction partners, previously characterized as RIP1 or RIP3 using transient expression systems (Upton et al., 2008), as well as the significance of RHIM-dependent signaling in antiviral host defense.
Figure 1. Role of MCMV M45 RHIM interactions in suppression of virus-induced death.
(A) IB analysis of vIRA expression from recombinant viruses to detect vIRA (M45), IE1, and βactin. NIH3T3 fibroblasts were either mock- or virus-infected at a multiplicity of infection (MOI) of 10 with bacmid-derived K181 (WT) or vIRA mutant virus (M45mutRHIM). An asterisk (*) designates a second independent WT or mutant virus isolate. (B) Replication levels and pathogenesis of WT and M45mutRHIM virus in severely immunodeficient NSG mice. At 16 d post inoculation (106 pfu into footpads), spleen (left panel), liver (middle panel), and salivary glands (right panel) were harvested from euthanized mice and viral titers determined by plaque assay. Each symbol represents one animal, solid horizontal lines represent the mean for each group, and dashed lines indicate the limit of detection in each assay. (C) Kaplan-Meier survival plot of NSG mice monitored for 42 days post infection with WT and M45mutRHIM virus (106 pfu into footpads). (D) Replication levels of WT and M45mutRHIM virus in spleen at 3 d post inoculation (106 pfu into peritoneal cavity) of BALB/c mice. (E) Replication levels of WT and M45mutRHIM virus in infected NIH3T3 fibroblasts (left panel) and SVEC4-10 endothelial cells (right panel) (MOI=10). Viral titers were determined by plaque assay at the indicated times post infection. (F) Viability of NIH3T3 fibroblasts (left panel) or SVEC4-10 cells (right panel) following infection with WT and M45mutRHIM virus. Where noted, zVAD-fmk (50 μM) was included throughout infection. Cell viability was determined by measuring intracellular ATP levels with a Cell Titer-Glo Luminescent Cell Viability Assay kit. Treatment with TNFα (25 ng/mL) and cycloheximide (CHX; 5 μg/mL) was used to induce apoptosis in uninfected NIH3T3 cells as a control. Error bars indicate standard deviation (SD) of the mean. See also the related Figure S1.
We next sought to investigate the importance of vIRA RHIM-dependent interactions in cell tropism, cell-specific viral replication and induction of cell death. Consistent with previous reports using vIRA null viral mutants (Brune et al., 2001; Mack et al., 2008; Upton et al., 2008), M45mutRHIM virus was severely attenuated for growth in SVEC4-10 cells (Figure 1E) which died within 18 h after exposure to mutant (Figure 1F, right panel). WT virus replicated to levels that were similar to NIH3T3 cells and had no impact on cell viability. NIH3T3 fibroblasts retained sensitivity to TNFα-induced apoptosis (when not infected with MCMV). As expected from previous investigations (Mack et al., 2008), mutant virus-induced cell death was unaffected by addition of the broad-spectrum caspase inhibitor, zVAD-fmk (Figure 1F, right panel). Additionally, the phenotype of two independently derived M45mutRHIM isolates was comparable (Figure S2A), confirming that the tetra-alanine mutation in M45 was responsible for the phenotype. Thus, the behavior of RHIM mutant virus recapitulates the behavior of previously characterized M45-deficient viruses, and clearly demonstrated that RHIM-dependent interactions are integral to vIRA cell death suppression. These data are consistent with our previously proposed model (Upton et al., 2008) where RHIM-dependent interaction(s) with RIP3 and/or RIP1 controls the antiviral cell death pathway triggered shortly after viral infection. Thus, MCMV induces programmed caspase-independent death in susceptible cells and virus-encoded vIRA naturally suppresses this pathway by interfering with RHIM-dependent pathways.
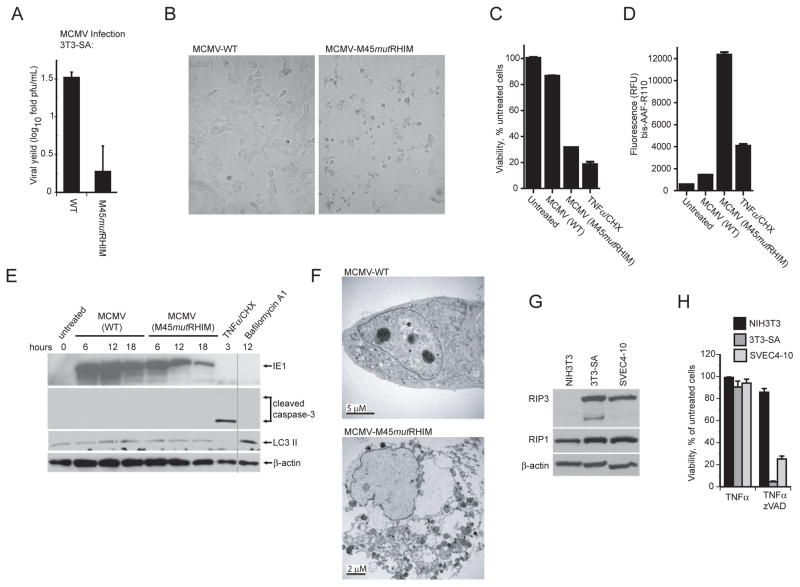
Given the proposal that vIRA represents an endothelial growth determinant (Brune et al., 2001), we were surprised when M45mutRHIM virus failed to replicate efficiently (Figure 2A) and, instead, induced premature death in 3T3-Swiss albino (3T3-SA) cells, another immortal fibroblast line that is fully permissive for WT MCMV (Figure S2B and data not shown). Exposure to mutant virus resulted in premature death similar to what was observed using SVEC4-10 cells, based on three independent lines of evidence: morphological evaluation (Figure 2B and Figure S2), intracellular ATP levels (Figure 2C) and release of intracellular proteases (Figure 2D). Multiple isolates of M45mutRHIM induced death on SVEC4-10 and 3T3-SA cells (Figure S2A). Like SVEC4-10 cells (Brune et al., 2001), 3T3-SA fibroblasts were fully susceptible to viral infection as assessed by expression of viral antigens (Figure S2B). Death of mutant virus infected cells was first detected between 5 and 9 hpi and approached maximal levels by 18 hpi, accompanied by cytoplasmic swelling, cell detachment, propidium iodide inclusion, and membrane rupture (Figure S2C, data not shown). This morphological evidence is most consistent with programmed necrosis (Festjens et al., 2006). To demonstrate that collapse in ATP levels (Figure 2C) was due to necrotic death rather than viral modulation of cellular respiration, we assessed the release of intracellular proteases into the culture medium. The dramatic increase in released proteases (Figure 2D) was consistent with dramatic loss of plasma membrane integrity associated with necrotic death, rather than other changes in cellular physiology during viral infection. In contrast to the virus-induced necrotic death, TNFα plus CHX was employed to induce apoptosis, and ATP levels decreased without a dramatic rise in extracellular protease activity. To assure that the virus induced release of proteases occurred independent of apoptosis or autophagy, appropriate markers were assessed by IB analysis of infected 3T3-SA cells. Infected cells did not exhibit any evidence of caspase-3 activation at any time from 6 through 18 h post infection (hpi) with mutant virus (Figure 2E). In addition, the levels of LC3 II remained similar in uninfected or cells infected with mutant or WT viruses, compared to Bafilomycin A1-treated controls. Ultrastructural analysis by electron microscopy (EM) failed to reveal evidence of membrane blebbing, nuclear condensation, or autophagosome formation in dying, mutant-infected cells compared to WT virus infection (Figure 2F and data not shown). Taken together with the caspase-independence, these observations indicate that the cell death pathway induced by mutant virus infection was unrelated to apoptosis or autophagy.
Figure 2. Cells sensitive to MCMV-associated programmed necrosis express high levels of RIP3.
(A) Viral yields determined by plaque assay at 72 hpi in 3T3-SA cells following infection with WT or M45mutRHIM virus (MOI=10). (B) Bright field micrographs of 3T3-SA cells infected for 18 h with either WT (left panel) or M45mutRHIM (right panel) virus. Magnification, 10X. (C) Viability of 3T3-SA cells assessed by intracellular ATP levels following infection with WT or M45mutRHIM virus. (D) Release of protease to the medium assessed in 3T3-SA cells using a CytoTox-Fluor kit. (E) IB of 3T3-SA cells infected with WT or M45mutRHIM virus (MOI=10) for the indicated times followed by detection of IE1, caspase-3, LC3 II and β-actin. Vertical line shows where lanes from the original gel were brought adjacent. In C - E, uninfected cells were treated to induce apoptosis (TNFα/CHX) as described in Figure 1F or with Bafilomycin A1 (250 nM) to induce LC3 II, or left untreated. (F) Transmission electron micrographs of 3T3-SA cells at 18 hpi with WT (top panel) or M45mutRHIM (bottom panel) virus. Size bars are indicated. (G) IB analysis of NIH3T3, 3T3-SA, and SVEC4-10 cells to detect RIP1, RIP3, and β-actin. (H) Viability of NIH3T3, 3T3-SA, and SVEC4-10 cells treated for 18 h with TNFα (25 ng/mL) in the absence or presence of zVAD-fmk (25 μM). See also the related Figure S2.
Given that recent studies have implicated high levels of RIP3 as a determinant in DR-induced necroptosis (Cho et al., 2009; He et al., 2009; Zhang et al., 2009), we next evaluated levels of the known vIRA interacting proteins, RIP1 and RIP3, in cells that were susceptible to virus-induced death (3T3-SA, and SVEC4-10) as well as in control NIH3T3 cells. IB analysis revealed that RIP3 levels were elevated in susceptible cells compared to control cells (Figure 2G); whereas, RIP1 levels were similar in all cell lines. SVEC4-10 and 3T3-SA cells were shown to be sensitive to TNFα and zVAD-fmk necroptosis (Figure 2H), paralleling susceptibility to virus-induced programmed necrosis. Together, these results suggested that, as is the case reported for DR-induced necroptosis (He et al., 2009; Zhang et al., 2009), RIP3 expression is a common property of cells sensitive to virus-induced necrosis, which serves to clarify and add mechanistic insight to prior observations regarding the premature death of SVEC4-10 cells to infection with vIRA-deficient virus (Brune et al., 2001), as well as to evidence that the pathway is caspase-independent (Mack et al., 2008). Thus, high levels of RIP3 are the defining characteristic of cells that are sensitive to either virus-induced or DR-induced necrotic death, implicating this adaptor in necrosis induced by different stimuli.
vIRA suppresses necroptosis in a RHIM-dependent fashion
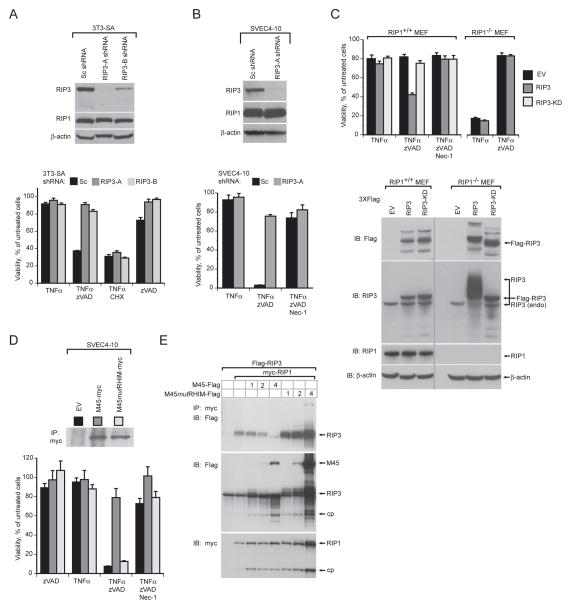
To establish the contribution of RIP1 and RIP3 to DR-induced necroptosis in SVEC4-10 and 3T3-SA cells, we employed specific shRNAs (RIP3-A and RIP3-B) to reduce expression levels of RIP3. This RNAi approach showed that RIP3 played a critical role in TNFα plus zVAD-fmk-induced necroptosis (Figure 3A and B). Addition of the RIP1-specific kinase inhibitor Nec-1 restored cell viability, consistent with prior studies (Degterev et al., 2008; Degterev et al., 2005) showing necroptosis to be RIP1 kinase-dependent (Figure 3B, bottom panel, Figure S3E, F, right panels). The contribution of RIP3 and RIP1 to necroptosis was further evaluated in RIP1−/− and control RIP1+/+ mouse embryonic fibroblasts (MEFs). RIP1−/− MEFs were insensitive to TNFα plus zVAD-fmk-induced necroptosis (Figure 3C, top panel). However, as expected (Kelliher et al., 1998; Wong et al., 2009), RIP1-deficient cells exhibited an increased sensitivity to TNFα–induced, caspase-dependent, apoptosis (Figure 3C, upper panel). IB analysis of EV transduced RIP1−/− and control MEFs showed similar levels of expression of endogenous RIP3 (Figure 3C, lower panel). To evaluate the impact of increasing RIP3 in the absence of RIP1, RIP1−/− and control MEFs were transduced with epitope-tagged WT or kinase-deficient (KD) RIP3 (Figure 3C, lower panel). Elevated levels of WT, but not KD, RIP3 conferred increased sensitivity of control MEFs to TNFα plus zVAD-fmk induced necroptosis (Figure 3C, upper panel) and this was reversed by Nec-1, showing a dependence on both RIP1 and RIP3 kinase activity. In contrast, overexpression of RIP3 in RIP1−/− MEFs failed to enhance susceptibility to either necroptosis or TNFα-induced apoptosis. Interestingly, elevated expression of RIP3, but not RIP3-KD, in RIP1−/− MEFs resulted in a dramatic RIP3 mobility shift, suggesting RIP1 may suppress a RIP3-kinase dependent post-translational modification of RIP3 in unstimulated cells (Figure 3C, lower panel). Taken together, these results indicate that RIP1 plays an essential role in induction of DR-associated necrotic death, that elevated levels of RIP3 are necessary for this pathway, and that overexpression of RIP3 does not overcome the requirement for RIP1 in DR-induced cell death. Furthermore, these data establishes the requirement for the protein kinase activity of both RIP1 and RIP3 in necrotic death induced following DR activation in sensitive cells. These independently derived results reaffirm the essential role of RIP3 and RIP1 in formation of an active RIP3-RIP1 kinase complex as well as the importance of their kinase domains to drive DR-induced necroptosis (Cho et al., 2009; He et al., 2009; Zhang et al., 2009).
Figure 3. MCMV RHIM-dependent suppression of RIP3-mediated necrosis.
(A) IB analysis (top panel) to detect RIP3, RIP1, and β-actin, and viability (lower panel) of 3T3-SA fibroblasts expressing a scramble control (Sc) or one of two RIP3-specific shRNAs (RIP3-A and RIP3-B) and treated to induce necroptosis as described in Figure 2H or to induce apoptosis as described in Figure 1F. (B) IB analysis (top panel) to detect RIP1, RIP3, and β-actin and viability (bottom panel) of SVEC4-10 cells expressing Sc or RIP3-A shRNA. Cells were treated to induce necroptosis as described in Figure 2H in the absence or presence of Nec-1 (30 μM). (C) Viability (top panel) and IB analysis to detect RIP1, RIP3, β–actin and Flag-epitope tagged proteins (lower panel) in RIP1+/+ and RIP1−/− MEFs transduced with empty vector (EV), Flag-tagged RIP3 (RIP3), or Flag-tagged kinase-deficient RIP3 (RIP3-KD) retroviral constructs. Transduced cells were treated as described in (B). The vertical line shows where lanes from the original gel were brought adjacent. (D) IB of myc-tagged proteins following IP with anti-myc conjugated agarose (top panel) of SVEC4-10 cells transduced with EV, M45-myc, or M45mutRHIM-myc retroviral constructs and viability (bottom panel) of these cells after treatment as described in (B). (E) IB of 293T cells transfected with Flag-tagged RIP3, myc-tagged RIP1 and 1, 2, or 4 μg of either M45-Flag or M45mutRHIM-Flag expression plasmids after IP with anti-myc agarose beads. In all samples, the amount of transfected DNA was held constant with addition of control plasmid DNA. IB detection was with anti-Flag or anti-c-Myc (9E10) antibody. Major protein species are identified, with “cp” indicating a processed form of RIP1 or RIP3. Error bars indicate SD of the mean.
Previously, vIRA was shown to suppress caspase-independent programmed cell death (Mack et al., 2008). To further investigate the role of vIRA in suppression of necroptosis and determine the contribution of RHIM-mediated interactions, SVEC4-10 cell lines stably expressing epitope-tagged WT (M45-myc) or mutant (M45mutRHIM-myc) vIRA were generated (Figure 3D, top panel) and compared to cells carrying an EV as control. As expected, expression of WT vIRA conferred protection to treatment with TNFα plus zVAD-fmk (Figure 3D, bottom panel) (Mack et al., 2008). Cells expressing M45mutRHIM-myc were as sensitive to death as EV control cells, indicating that vIRA is a potent RHIM-dependent suppressor of DR-induced necroptosis.
Given that both RIP1 and RIP3 contributed to the necroptosis pathway in the expected fashion (Cho et al., 2009; He et al., 2009, Zhang, 2009 #340), we sought to evaluate the potential role of vIRA as a competitor of RIP1-RIP3 complex formation. FLAG-RIP3 and myc-RIP1 were coexpressed in 293T cells in the presence of increasing amounts of either WT M45-FLAG or mutant M45mutRHIM-FLAG. IP followed by IB analysis revealed a RHIM-dependent vIRA-mediated inhibition of RIP1-RIP3 complex formation (Figure 3E). This demonstration provides mechanistic insight into how vIRA suppresses the execution of necroptosis initiated by DR- signaling but leaves open whether RIP1, RIP3 or both are directly targeted by the viral cell death suppressor.
MCMV-induced programmed necrosis is RIP3-dependent, but RIP1-independent
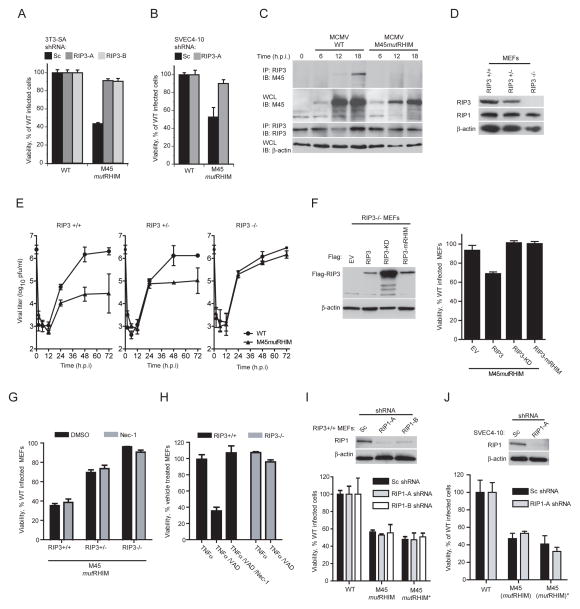
Having shown that vIRA suppresses necroptosis, which requires the expression and kinase activity of RIP3 and RIP1 as well as a RIP3-RIP1 complex (Cho et al., 2009; He et al., 2009; Zhang et al., 2009), we characterized the cellular requirements for MCMV-associated programmed necrosis. RIP3-knockdown rendered either 3T3-SA or SVEC4-10 cells resistant to programmed necrosis induced by mutant virus (Figure 4A and B), indicating a requirement for RIP3. To clarify the role of RIP3, control (RIP3+/+), heterozygous (RIP3+/−), and homozygous (RIP3−/−) deficient MEF cells were isolated, and assessed for virus replication and sensitivity to programmed necrosis. RIP3 levels were assessed by IB in cell lysates and shown to reflect gene copy (Figure 4D). WT MCMV replicated to comparable high titers in all MEF lines tested, and, as expected, M45mutRHIM virus was significantly attenuated in RIP3+/+ cells (Figure 4E, left panel) where RIP3 levels were the highest (Figure 4D). Mutant virus replication was fully rescued in RIP3−/− MEFs (Figure 4E, right panel). Consistent with a relationship between RIP3 levels and sensitivity to this death pathway (Figure 4D), replication levels and sensitivity to death were intermediate in RIP3+/− cells (Figure 4E, middle panel, and Figure 4G). Furthermore, RIP3+/+ MEFs died following mutant virus infection, whereas RIP3−/− cells survived, sustaining the evidence that RIP3 is required for virus-induced death to occur (Figure 4G). Here, RIP3 levels correlated with cellular sensitivity to virus-induced programmed necrosis as well as to the concomitant attenuation of viral growth. When RIP3−/− MEFs were reconstituted with WT, KD or mutant RHIM (mRHIM), RIP3 (Figure 4F, left panel) and infected with mutant virus, only WT RIP3 reconstituted cells exhibited increased sensitivity to virus-induced necrosis (Figure 4F, right panel). These data are consistent with a direct role of RIP3, as well as a requirement for an intact RIP3 kinase domain and RHIM for cell death signal transduction in response to viral infection.
Figure 4. RIP3 is required, and RIP1 is dispensable for MCMV-associated programmed necrosis.
(A) Viability of 3T3-SA cells expressing Sc, RIP3-A or RIP3-B shRNAs determined 18 hpi with WT or M45mutRHIM virus (MOI of 10). (B) Viability of SVEC4-10 cells using a subset of conditions described in (A). (C) IB of 3T3-SA cells infected with WT or M45mutRHIM virus (MOI of 5), harvested at indicated times for IP of RIP3 followed by detection of vIRA (M45) and RIP3. IB using ~5% of cell lysate to detect vIRA (M45) and β–actin. (D) IB of RIP3+/+, RIP3+/−, and RIP3−/− MEFs to detect RIP3, RIP1, and β–actin. (E) Replication of WT and M45mutRHIM viruses (MOI of 5) on RIP3+/+ (left panel), RIP3+/− (middle panel), and RIP3−/− (right panel) MEFs over a 72 h time course. Viral titers were determined by plaque assay with the first (0 h) time point representing the amount of virus in the inoculum. (F) IB analysis for FLAG-tagged proteins as well as β-actin (left panel) in RIP3−/− MEFs expressing FLAG-tagged RIP3, RIP3-KD, or RIP3-mRHIM and viability of reconstituted cells (right panel) infected with M45mutRHIM and WT virus. (G) Viability of RIP3+/+, RIP3+/−, and RIP3−/− MEFs infected with WT or M45mutRHIM virus in the presence or absence of Nec-1 (30 μM). (H) Viability of RIP3+/+ and RIP3−/− MEFs treated to induce necroptosis as described in Figure 2H in the presence or absence of Nec-1 (30 μM). (I) IB analysis for RIP1 as well as β–actin (top panel) and viability (bottom panel) of WT (RIP3+/+) MEFs stably expressing Sc, RIP1-A or RIP1-B shRNAs. Cell viability was determined for cells infected with WT or either of two independent isolates of M45mutRHIM virus. (J) IB analysis (top panel) and viability (bottom panel) of SVEC4-10 cells using a subset of conditions applied in (I). Error bars indicate SD of the mean. See also the related Figure S3.
We showed that vIRA bound RIP3 in a RHIM-dependent manner when both were transiently overexpressed (Upton et al., 2008). Here we have revealed a requirement for RIP3 in virus-associated programmed necrosis. To formally demonstrate that vIRA directly targets RIP3 to modulate this death process during infection, we infected necrosis-sensitive 3T3-SA cells. Co-IP of M45 protein from infected cell lysates at 12 and 18 h postinfection with antibody to RIP3 is consistent with a physical interaction of vIRA with endogenous RIP3 (Figure 4C). The absence of detectable vIRA in RIP3 immunoprecipitates from M45mutRHIM virus-infected cells is consistent with this being a RHIM-dependent interaction. We were unable to detect an interaction between endogenous RIP3 and RIP1 during viral infection with either WT or mutant viruses (data not shown). These data suggest that virus-associated programmed necrosis is distinct from DR-induced necroptosis because the pathway does not involve RIP1.
To more formally investigate the role of TNFα as well as RIP1 in virus-associated programmed necrosis, we sought to interfere with mediators and adaptors known to play roles in necroptosis. First, infected cells were treated with TNFα-neutralizing antibody over a range of effective concentrations (Figure S3D) without observing any impact on virus-induced necrosis in susceptible cell types (Figure S3A, B, C). Second, infected cells were treated with the RIP1-specific kinase inhibitor Nec-1 to determine whether RIP1 kinase activity contributes to virus-induced programmed necrosis. Consistent with the inability to detect interactions between RIP3 and RIP1 during infection, treatment with Nec-1 over a range of doses failed to restore viability of infected primary MEF, 3T3-SA, or SVEC4-10 cells (Figure 4G, grey bars; Figure S3E, F, G). In contrast, Nec-1 treatment as low as 15–30 μM was sufficient to suppress TNFα-induced necroptosis in these same cell lines (Figure 4G, S3E, F), confirming both the sensitivity of these cells to DR-induced necroptosis and efficacy of Nec-1 as a RIP1 inhibitor that does not impact RIP3. Thus, RIP1 kinase activity is dispensable for virus-associated programmed necrosis, in contrast to its critical role in necroptosis. Since SV40-transformed MEFs, including both the RIP1+/+ and RIP1−/− MEFs used in Figure 3C, were resistant to virus-induced cell death (data not shown), we knocked down RIP1 expression by RNAi in either primary MEFs or SVEC4-10 cells with multiple RIP1-specific hairpins (Figure 4I, J), and these cells failed to suppress virus-induced necrosis. This result was recapitulated using independently derived isolates of M45mutRHIM mutant virus (Figure 4I, J). RIP3 scores as necessary whereas RIP1 appeared dispensable by either knockdown or chemical inhibition in virus-induced programmed necrosis. Thus, unlike TNFα-induced necroptosis, virus-induced necrosis does not rely on a RIP1/RIP3 kinase complex and instead appears to be a novel RIP3-dependent programmed necrosis pathway.
Normalization of M45 mutant virus phenotype in RIP3-deficient mice
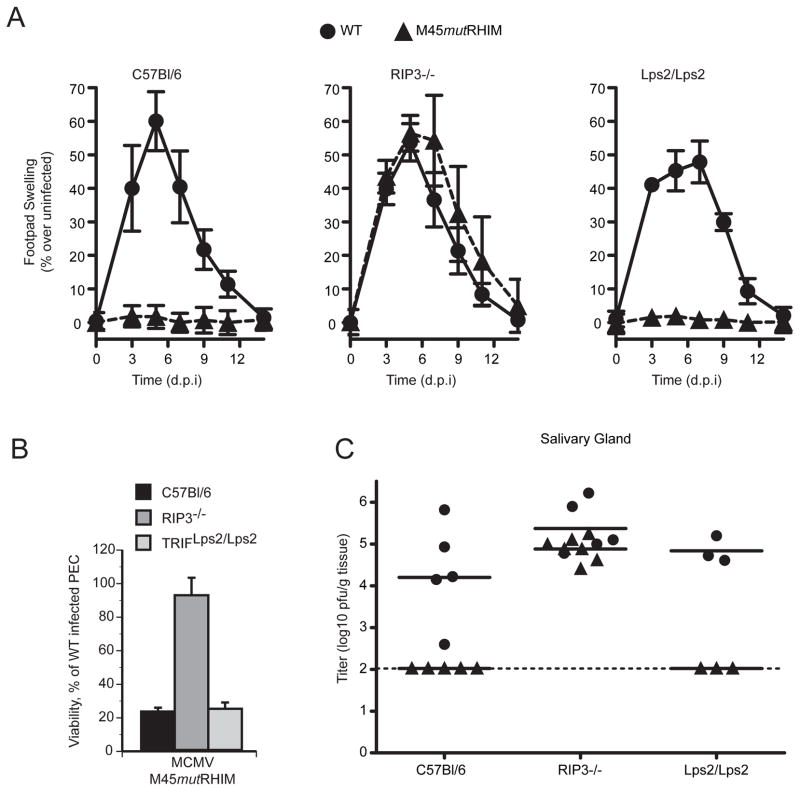
Given that vIRA RHIM-deficient mutant MCMV induces a form of RIP3-dependent programmed necrosis that strongly attenuates viral pathogenesis, we inoculated RIP3−/− as well as control RIP3+/+ C57BL/6 mice in order to follow initial inflammatory events induced by WT or mutant virus. Following footpad route of inoculation, WT virus replicates at the portal of entry, followed by dissemination to distal organs. This route allows for the assessment of virus-induced inflammation at the site of inoculation, which is influenced by the MCMV-encoded CC-chemokine homolog MCK-2 (Saederup et al., 2001). During infection, WT MCMV induces characteristic swelling of the footpad compared to uninfected controls, peaking at day 5 post-infection and resolving by 14 days. Following inoculation, M45mutRHIM virus infection induced swelling in RIP3−/− mice comparable to WT virus infection of either RIP3+/+ or RIP3−/− mice, reaching greater than 50% increase in size over uninfected paws (Figure 5A). In contrast, mutant virus did not induce significant swelling in RIP3+/+ mice, consistent with an attenuated phenotype. Thus, in the absence of RIP3 function, mutant virus infection recapitulated early inflammatory events characteristic of WT infection.
Figure 5. M45mutRHIM attenuation in vivo is specifically normalized in RIP3-deficient mice.
(A) Swelling induced by WT or M45mutRHIM virus infection of C57BL/6 (RIP3+/+), RIP3−/−, and TRIF-deficient (Lps2/Lps2) mice. Groups of five (C57BL/6 and RIP3−/−) or three (Lps2/Lps2) mice were inoculated (106 pfu) in footpads, and thickness was measured with a digital caliper (Saederup et al., 2001), and mean values were plotted at the indicated times over a 14 day time course. Error bars indicate standard error of the mean. (B) Viability of explanted, cultured RIP3−/−, Lps2/Lps2, or C57BL/6 (WT) PECs at 18 hpi with either WT or M45mutRHIM. Error bars indicate SD of the mean. (C) Salivary glands were harvested from euthanized mice (described in A) and titers determined by plaque assay. Each symbol represents one mouse, and solid horizontal lines represent the mean for each group. Dotted line is the limit of detection in this assay.
Given the importance of RHIM-mediated activity in suppressing virus-associated death as well as the observation that RIP3 associates with TRIF, we sought to determine whether this cellular RHIM adaptor (Kaiser and Offermann, 2005; Meylan et al., 2004) contributed to the inflammatory phenotype. TRIF is known to influence WT MCMV infection (Tabeta et al., 2004). Despite previous demonstration that vIRA is able to suppress RHIM-dependent cell death signaling relayed via TRIF (Upton et al., 2008), when inoculated in footpads, TRIF-mutant (TrifLps2/Lps2) mice (Hoebe et al., 2003) failed to exhibit any difference in behavior from WT mice (Figure 5A, right panel). Thus, vIRA plays a critical RIP3-dependent, TRIF-independent step in viral pathogenesis.
When explanted RIP3+/+, RIP3−/−, and TrifLps2/Lps2 peritoneal exudate cells (PEC) were infected ex vivo with either WT or mutant virus, only RIP3−/− PECs were resistant to mutant virus-induced programmed necrosis (Figure 5B), consistent with the behavior of primary fibroblasts to mutant virus infection. Given that PECs express TLR3 and TLR4, this experiment excluded any contribution of TRIF-dependent TLR-signaling to RIP3-dependent virus-induced cell death. The TLR adaptor TRIF does not contribute to virus-induced programmed necrosis.
Finally, we sought to evaluate the role of RIP3 in viral pathogenesis. Mice were inoculated in footpads and sacrificed 14 days post infection, when salivary glands were harvested and viral titers were determined by plaque assay. Salivary glands are the major site of secondary MCMV replication as well as the source of virus transmission between hosts. Virus levels in this organ remain a highly sensitive indicator of successful infection and dissemination to secondary organs in the intact host. Virus titers in salivary gland homogenates of RIP3−/− mice were comparable to WT virus (Figure 5C). The complete normalization of mutant virus in RIP3−/− mice provides formal proof that RIP3 is the relevant target of vIRA modulation under physiological conditions. In the absence of RIP3, vIRA function appears dispensable for normal WT levels of viral replication or dissemination. Consistent with the experiments in NSG and BALB/c mice, C57BL/6 mice were unable to support infection and dissemination of mutant virus and mutant virus was not able to disseminate to salivary glands of TRIF-defective mice (Figure 5C). WT MCMV disseminated at similar levels to salivary glands of control, RIP3−/− and TRIF-defective C57BL/6 mice. Normalization of mutant virus behavior by elimination of a host determinant provides compelling and unambiguous evidence that RIP3 is the target of vIRA-mediated modulation during MCMV infection, and further suggests that RHIM-dependent suppression of RIP3 function in virus-associated cell death by vIRA is essential for MCMV pathogenesis.
Discussion
Manipulation of host cytokine and cell death pathways affords pathogens the opportunity to maintain an environment necessary for efficient replication to establish a foothold within the host organism. In this work, which derives in part from the goal of elucidating mechanisms of cell death suppression by a natural pathogen of mice, we have demonstrated a critical role for RIP3 as a positive regulator of a cell-intrinsic antiviral programmed necrosis triggered during MCMV infection. Our results show a dependence on RIP3 function in both virus-associated programmed necrosis and DR-associated necroptosis. We also provide a clear mechanistic distinction between these two pathways. Whereas necroptosis is dependent on RIP1 and a RIP1-RIP3 signaling complex (Cho et al., 2009; He et al., 2009; Zhang et al., 2009), virus-induced death is dependent on RIP3 but not RIP1. MCMV encoded vIRA suppresses either of these necrotic death pathways in a RHIM-dependent fashion, suggesting that the common target of this viral cell death suppressor is RIP3. The dramatic differences in behavior of viruses expressing WT or RHIM-deficient form of vIRA, assayed in vitro and in vivo, demonstrate that this death suppressor is critical for MCMV replication and pathogenesis. The normalization of mutant virus behavior when RIP3 function is eliminated in the host formally establishes the biological significance of vIRA-RIP3 interactions. By revealing the commitment of a specific pathogen-encoded suppressor to necrotic death pathways, our data validates RIP3-dependent programmed necrosis as a central player in host defense, and reveal yet another cytomegalovirus immune escape mechanism contributing to viral pathogenesis.
The normalization of vIRA-deficient MCMV behavior in RIP3 deficient cells and mice provides unambiguous evidence that virus-induced inflammatory responses, viral replication and viral pathogenesis are all directly linked to RIP3-mediated events, and these most likely revolve around the two pathways that vIRA blocks: virus-associated programmed necrosis and DR-associated necroptosis. Together with the surprising demonstration the RHIM-mediated vIRA function is independent of RIP1 as well as TRIF, these data strongly argue that RIP3 is a dominant adaptor controlling cellular necrotic pathways in viral pathogenesis.
Restoration of an attenuated vIRA viral mutant phenotype in vivo by the elimination of the host intracellular adaptor such as RIP3 has rarely been achieved in studies on viral pathogenesis. The most relevant to our studies is the demonstration that herpes simplex virus ICPγ34.5 uniquely targets protein kinase R (Leib et al., 2000), an RNA binding adaptor protein that shuts down translation in virus-infected cells. The attenuation of ICPγ34.5 mutant virus was completely normalized in protein kinase R-deficient mice. This strategy has also been employed to show the in vivo function of a number of herpesvirus immune evasion genes acting on the immune system. For example, MCMV m157 and m04 secreted gene products target the natural killer cell receptors Ly49H (Bubic et al., 2004) and Ly49P (Kielczewska et al., 2009), respectively, and murine gammaherpesvirus 68 regulator of complement activation (Kapadia et al., 2002) as well as herpes simplex virus glycoprotein C (Friedman et al., 1996; Lubinski et al., 1999; Lubinski et al., 1998) directly target complement factor C3 in pathogenesis. Here, we provide in vivo evidence that MCMV M45 gene product vIRA mediates its biological function by antagonizing the host necrosis-regulating kinase RIP3, and, furthermore, that the RIP3-dependent death pathway, as well as vIRA suppression, rely on RHIM-mediated interactions. Thus, our demonstration that virus carrying RHIM-deficient vIRA exhibited normalized replication in RIP3-deficient mice together with the evidence that restoration of WT RIP3 expression, but not KD or RHIM-deficient RIP3, sensitizes RIP3-deficient cells to virus-induced programmed necrosis, demonstrates the exclusive interplay of RIP3 and vIRA in MCMV pathogenesis.
Virus- and DR-associated programmed necrosis share a requirement for RIP3 kinase activity and RHIM-dependent interactions (Cho et al., 2009; Hitomi et al., 2008; Zhang et al., 2009). High levels of RIP3 remain the common feature that predicts sensitivity to necrotic death from either inducer. In addition, endothelial cell type (Brune et al., 2001), per se plays no role in susceptibility to premature death induced by vIRA mutant virus. The recent findings that high RIP3 levels confer sensitivity of human and murine cell lines to TNFα-induced necroptosis (He et al., 2009; Zhang et al., 2009), are entirely consistent with the results presented here. One characteristic of DR-induced necroptosis, the requirement for kinase activity and RHIM-dependent signaling of RIP1 (Christofferson and Yuan, 2009; Declercq et al., 2009; Festjens et al., 2007; Galluzzi and Kroemer, 2008) distinguishes this necrotic pathway from virus-induced necrosis. Nec-1 treatment inhibits the formation of a RIP1-RIP3 complex, RIP3 activation, and cell death in response to TNFα (Cho et al., 2009; He et al., 2009). Treatment with this RIP1-specific inhibitor has no impact on RIP3 activation or signaling in virus-induced necrosis. Overexpression of RIP3 has previously been reported to induce cell death (Kasof et al., 2000; Pazdernik et al., 1999; Sun et al., 1999; Yu et al., 1999), including in situations where RIP1 expression has been eliminated (Zhang et al., 2009). While this work indicated RIP3 alone may be sufficient to drive necrotic death, the data reported here show physiological relevance of a RIP3-dependent, RIP1 independent pathway. The cell death pathway induced by virus infection is naturally executed in cells that can also support DR-induced necroptosis. The RIP1 as well as TRIF-independence of virus-induced necrosis, raises a need to identify additional RHIM-dependent interactions of RIP3 as well as other downstream events in RIP3-dependent, RIP1-independent programmed necrosis.
vIRA likely triggers a range of events when complexed with RIP3 that may contribute to suppression of virus- or DR-associated necrosis. vIRA is composed of two distinct regions, a 277 amino terminal RHIM-containing region is sufficient for suppression of cell death (Upton et al., 2008) and a 801 aa region that contains the herpesvirus-conserved ribonucleotide reductase large subunit homology that has been assigned conflicting roles in suppression (Mack et al., 2008) and activation of NF-κB expression (Rebsamen et al., 2009). The direct interaction of vIRA with RIP3 opens the way for other activities of vIRA to potentially contribute to parallel signaling pathways. Regardless of how downstream signaling proceeds, it is important to recognize that neither the carboxyl-terminus nor other regions of vIRA can mediate suppression of virally induced programmed necrosis when RHIM-dependent interactions are disrupted.
The cellular death response to infection with vIRA-deficient MCMV was initially characterized as apoptosis (Brune et al., 2001) with caspase-independence only recognized recently (Mack et al., 2008). Here, we have formally demonstrated that a four amino acid substitution to prevent RHIM-dependent interactions recapitulates the expected behavior of vIRA-null mutant phenotype (Brune et al., 2001), and conclusively show, by morphological, biochemical, and cell biological criteria, that the cell death pathway is RIP3-dependent programmed necrosis. Molluscum contagiosum, equine herpesvirus-2 and Kaposi’s sarcoma-associated herpesvirus encode proteins that can suppress DR-induced necroptosis (Chan et al., 2003), although the mechanisms by which these viral genes suppress necroptosis, and their specific role(s) in pathogenesis, remain undetermined. Comparison to investigations on the RIP3-dependence of vaccinia-induced liver pathology, inflammation and viral replication in mice (Cho et al., 2009) reveals potential parallels and differences. The behavior of vaccina has been interpreted to be a consequence of TNFα production, and supports findings that cytokine responses are a primary mechanism used to control poxviruses during zoonotic infection (Wang et al., 2008). While DR signaling (Benedict, 2003; Benedict et al., 2003) as well as TLR signaling (Hoebe et al., 2003; Tabeta et al., 2004) contribute to the replication levels of WT MCMV in mice, we found no evidence that either is involved in initiating MCMV-associated programmed necrosis during infection. We speculate that this antiviral death pathway is a cell intrinsic response to viral infection that responds to the initial events following entry into cells.
While we have identified RIP3 as the executioner of MCMV-induced programmed necrosis, the cellular processes leading to activation of this pathway by virus remain unidentified. UV-inactivated mutant virus does not induce cell death (Brune et al., 2001), and preliminary evaluation suggests that inhibition of viral DNA replication fails to protect from programmed necrosis. Thus, the RIP3-activating step likely originates from early events in virus-infected cells perhaps through cellular processes altered or usurped by viral infection. Alternatively, sensing of invading pathogens by PRRs initiate signals to contend with pathogens, and several of these sensor proteins are known to recruit RIP3 (Kaiser et al., 2008; Kumar et al., 2009; Meylan et al., 2004; Palm and Medzhitov, 2009; Saito and Gale, 2007). TLR3-TRIF contributes to the innate immune response to MCMV infection, though less significantly than TLR9 (Hoebe et al., 2003; Tabeta et al., 2004), and RIP3 binds to TRIF in a RHIM-dependent interaction (Kaiser and Offermann, 2005; Meylan et al., 2004), although the significance of this interaction remains unclear. The capacity of vIRA to suppress RHIM-dependent death signals relayed via TRIF (Upton et al., 2008) was not sustained here as TRIF-deficient peritoneal macrophages remained susceptible to killing by mutant virus and mice lacking TRIF were no more susceptible to mutant virus than WT mice. Thus suppression of programmed necrosis by vIRA during infection is unlikely to be tied to other characterized RHIM-dependent pathways dependent upon TRIF. Our results raise the possibility that another pathogen sensor pathway drives RIP3 activation, and the IFN-inducible, cytosolic DNA sensor DAI/ZBP1/DLM-1 (Takaoka et al., 2007) remains an attractive candidate to fulfill such a role, since it contains a RHIM and physically interacts with RIP3 as well as vIRA (Rebsamen et al., 2009). Defining the cellular processes required for induction of RIP3-dependent death during MCMV infections remains a significant future line of investigation.
Cytomegaloviruses encode an array of cell death suppressors to counter apoptotic cell death pathways that may dominate in different cell and tissue settings (McCormick, 2008). MCMV encodes three genes that target core components of the apoptotic machinery: viral mitochondrial inhibitor or apoptosis, vMIA (Goldmacher et al., 1999; McCormick et al., 2005), viral inhibitor of Bak oligomerization, vIBO (Cam et al., 2009), and viral inhibitor of caspase-8 activation, vICA (Skaletskaya et al., 2001). Together, these functions antagonize caspase-dependent cell death pathways that would otherwise compromise viral replication (Cicin-Sain et al., 2008; Menard et al., 2003). vMIA and vIBO function at the level of the mitochondria to suppress amplification of pro-apoptotic signals induced by extrinsic and intrinsic stimuli (Cam et al., 2009; Goldmacher et al., 1999; McCormick et al., 2005). vICA inhibits caspase-8 activation and prevents DR-induced apoptosis (McCormick et al., 2005; Skaletskaya et al., 2001), and plays a critical role supporting viral replication in macrophages (Menard et al., 2003); McCormick et al, manuscript in revision). vICA suppression of apoptosis is critical in vivo, where the defect is restored by expression of FADD-DN (Cicin-Sain et al., 2008). The pro-necrotic activity of RIP3 is inactivated by caspase-8 processing (Feng et al., 2007), suggesting that, in instances of infection by pathogens encoding caspase-8 inhibitors such as vICA, a virally encoded inhibitor of programmed necrosis, like vIRA, prevents the induction of necrosis. A recombinant viral mutant lacking the Bax inhibitor, vMIA, replicates comparable to WT in visceral organs of infected animals, but is attenuated for leukocyte-dependent dissemination of virus to the salivary gland (Manzur et al., 2009). Mutants lacking the caspase-8 inhibitor vICA are attenuated in vivo following systemic inoculation, and are severely attenuated for dissemination to salivary glands (Cicin-Sain et al., 2005; Cicin-Sain et al., 2008). These results have supported the importance of modulating host apoptotic pathways in MCMV pathogenesis. It is interesting to note that the attenuation of vIRA mutants in vivo is tied to suppression of RIP3 activation and is as critical to viral pathogenesis as the suppression of Bax activation or caspase-8 activation, by vMIA or vICA, respectively.
Conclusions
The identification of vIRA as an inhibitor of RIP3-dependent programmed necrosis in a natural biological setting extends the role of RIP3 as a vital player in host antiviral defense, and underscores the critical role of vIRA RHIM-dependent activities in suppression of programmed necrosis in MCMV pathogenesis. This study also begins to illustrate the balance of necrotic and apoptotic pathways in host defense against viruses, supporting the growing evidence that viruses exploit multiple innate immune processes to influence a wide range of alternative immune effector activities.
Materials and Methods
Reagents
CHX, Bafilomycin A1, and dimethyl sulfoxide (DMSO) were obtained from Sigma Aldrich. Nec-1 and z-VAD-fmk were purchased from Calbiochem and Enzo Life Sciences, respectively. Recombinant mouse TNFα was from PeproTech and anti-TNFα antibody was from R&D Systems. The following antibodies were used for IB analysis: mouse anti-β–actin (clone AC-74; Sigma), mouse anti-RIP1 (clone 38; BD Biosciences), rabbit anti-RIP3 (Imgenex), rabbit anti-cleaved caspase-3 (Cell Signaling Technology), rabbit anti-LC3B (Cell Signaling Technology), mouse anti-FLAG (M2 clone) peroxidase conjugate (Sigma Aldrich), mouse anti-c-Myc (clone 9E10, Sigma Aldrich), rabbit anti-c-Myc peroxidase conjugate (Sigma Aldrich), anti-mouse IgG-HRP (Vector Laboratories), anti-rabbit IgG-HRP (Vector Laboratories), anti-MCMV M45/vIRA (gift from David Lembo, University of Turin), and anti-MCMV IE1 (gift from Stipan Jonjic, University of Rijeka). Immunoprecipitations were performed with rabbit anti-c-Myc agarose conjugate (Sigma Aldrich) or goat anti-RIP3 (clone C-16; Santa Cruz Biotechnology) and Protein A/G agarose (Santa Cruz Biotechnology).
BAC mutagenesis and recombinant viruses
Plasmid pSIM6 encoding lambda red recombination functions (Datta et al., 2006) was introduced into bacteria carrying a bacterial artificial chromosome (BAC) harboring the MCMV K181(Perth) strain genome (Redwood et al., 2005). Recombineering for K181-BAC mutagenesis and diagnostics was performed essentially as previously described (Tandon and Mocarski, 2008), and is outlined in Supplemental Materials. The supernantant of NIH3T3 cells transfected with WT and mutant K181-BAC DNA was collected and M45mutRHIM viruses were identified by replication on NIH3T3 cells (ATCC number CRL-1658) and purified as previously described (Redwood et al., 2005). WT and mutant viruses were propagated and titered by plaque assay on NIH3T3 cells as previously described (Saederup et al., 2001). Medium from infected cultures was clarified by centrifugation at 2500 g and virus collected by sedimentation at 17,000 g and stored at −80°C until use. Single step growth curves and viral yield assays were performed by infecting cell lines in 6 well plates at the indicated MOI in a volume of 0.5 ml for 2 hours at 37°C with periodic rocking. Following adsorption, inoculums were removed, cells washed three times with PBS, and refed with complete media. Samples were harvested at the indicated times, and virus growth assessed by plaque assay.
Mice, infections, and organ harvests
C57BL/6, BALB/c, TRIF mutant (Strain - C57BL/6J-Ticam1Lps2) (Hoebe et al., 2003), and NSG mutant (NOD.cg-PrkdcscidIl2rgtm1Wjl/Sz) (Shultz et al., 2005) mice were obtained from Jackson Laboratory. RIP3−/− mice (Newton et al., 2004) were provided by Francis Chan (University of Massachusetts) with permission from Vishva Dixit (Genentech). 8- to 12-week-old mice were inoculated with 106 pfu into a rear footpad or by intraperitoneal injection as previously described (Saederup et al., 1999). Upon sacrifice, organs were removed using aseptic technique, placed in 1 ml complete DMEM and frozen at −80°C. Organs to be titered were thawed, disrupted by sonication and viral growth assessed by plaque assay. Resident peritoneal macrophages were collected from euthanized animals by DMEM lavage, resuspended in DMEM containing 10% FBS and seeded at a density of 5 × 104 cells per a well in a 96 well plate 18 h prior to infection. For virulence studies, animals losing over 20% body weight or displaying severe signs of CMV-disease (ruffled fur, hunched posture, dehydration, diminished responsiveness) were scored and euthanized. Mice were bred and maintained by Emory University Division of Animal Resources in accordance with Institutional Guidelines, and all procedures were approved by the Emory University Institutional Animal Care and Use Committee.
Cell culture and embryonic fibroblast isolation
NIH3T3 fibroblasts, 3T3-SA (ATCC number CCL-92), SVEC4-10 (ATCC CRL-2181), HEK293T, L929, and MEFs were all maintained in DMEM containing 4.5 g/mL glucose, 10% FBS (Atlanta Biologicals), 2 mM L-glutamine, 100 U/mL penicillin, and 100 U/mL streptomycin (Invitrogen). SV40T transformed RIP1−/− and control WT MEFs (Wong et al., 2009) were kindly provided by John Silke (LaTrobe University) with permission from Michelle Kelliher (University of Massachusetts). Murine embryonic fibroblasts were isolated from timed pregnancies at day E14.5–16.5, as previously described (Pollock and Virgin, 1995).
Immunoblot and Immunoprecipitations
Immunoprecipitation were performed essentially as described previously (Kaiser and Offermann, 2005), with minor modifications. Clarified cell lysates were incubated overnight with 25 μl of anti-c-Myc agarose conjugate slurry or 1 μg of goat-anti-RIP3 antibody and 25 μl of Protein A/G slurry. Immunoprecipiates were washed 4–6 time prior to analyis. Cell lysates and immunoprecipitated samples were separated on Criterion gels (Bio-Rad), transferred to Immobilon PVDF membranes (Millipore), and subjected to IB analysis as previously described (Kaiser and Offermann, 2005). Detection of c-myc- and RIP3- immunoprecipitates were performed with mouse anti-c-myc (clone 9E10) or rabbit anti-RIP3 antibodies, respectively.
Plasmids, Transfections, and Transductions
Transfections were performed using Lipofectamine 2000 (Invitrogen) and DNA at a 1:1 ratio diluted in Opti-MEM (Invitrogen). Carboxyl-terminal Flag-epitope tagged WT and mutRHIM M45 were generated by subcloning the respective ORF into pCMV-TAG4A (Stratagene). pCMV10-3xFLAG-RIP3 and pcDNA3-6myc-RIP1 (Kaiser and Offermann, 2005) have been previously described. Flag-RIP3, Flag-RIP3-KD (Kaiser et al., 2008), and Flag-RIP3-mRHIM (Kaiser and Offermann, 2005), as well as M45-myc, and M45mutRHIM-myc (Upton et al., 2008) were subcloned into the pQCXIH (Clontech) retroviral construct. The pLKO.1-based RIP3-A (TRCN0000022535), RIP3-B (TRCN0000022538), RIP1-A (TRCN0000022467), and RIP1-B (TRCN0000022464) shRNA constructs were obtained from Open Biosystems. The pLKO.1-Scramble control shRNA vector (Sarbassov et al., 2005), as well as lentiviral and retroviral production, infection, and selection have all been described (Kaiser et al., 2008).
Cell Viability Assays
Cells (5000 cells/well) were seeded into 96-well plates. 16–18 hours post-seeding, media was replaced with 50 μl of viral inoculum containing 10 pfu/cell (MOI=10.0). Nec-1 at the indicated concentration was added 1 h prior to infection, and maintained for the duration of the assay. Inoculums were removed 2 h post infection, and replaced with 50 μl of complete media. Alternatively, cells were treated with the indicated concentrations of TNFα, CHX, zVAD-fmk and/or Nec-1. The Nec-1 vehicle, DMSO, was held constant for all cells. Viability was determined 18 h post infection/treatment. Unless otherwise indicated, the viable cells per a well was determined indirectly by measuring the intracellular levels of ATP as a marker for cell survival using the Cell Titer-Glo Luminescent Cell Viability Assay kit (Promega) according to the manufacturer’s instructions. 96-well plate assays were allowed to reach RT prior to adding 50 μL of reagent to each well and then shaken for 10 minutes. Released protease activity was measured using the CytoTox-Fluor Assay kit (Promega) according to the manufacturer’s instructions. Briefly, 50 μl of assay reagent was added to each sample well and incubated at room temperature for 2 h. Luminescence or fluorescence was measured on a Synergy HT Multi-Detection Microplate Reader (Bio-Tek).
Microscopy
3T3-SA cells were infected with WT or M45mutRHIM viruses (MOI of 10) for the indicated time in 6- or 12-well dishes, and bright field images were acquired on an AxioCam MRC5 camera attached to a Zeiss Axio Imager A1 and processed with AxioVision Release 4.5 software. Samples for EM were prepared by infecting 3T3-SA cells with WT or M45mutRHIM viruses (MOI of 10) for 18 h. Monolayer cells were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2) for 2 h at 4°C. Cells then were washed with the same buffer and postfixed with buffered 1.0% osmium tetroxide at room temperature for 1 h. Following several washes with 0.1 M cacodylate buffer, cells were dehydrated with ethanol, infiltrated, and embedded in Eponate 12 resin (Ted Pella Inc.). Ultrathin sections (60 to 70 nm) of monolayer cells were cut and counterstained using uranyl acetate and lead citrate. The examination of ultrathin sections was carried out on a Hitachi H-7500 transmission electron microscope.
Supplementary Material
Acknowledgments
We thank Vishva Dixit and Kim Newton (Genentech) for RIP3−/− mice, Francis Chan (University of Massachusetts) for facilitating transfer of RIP3−/− mice, John Silke (LaTrobe University) and Michelle Kelliher (University of Massachusetts) for RIP1−/− MEFs, David Lembo (University of Turin) for M45/vIRA antibody, and Stipan Jonjic (University of Rijeka) for MCMV IE1 antibody. We also thank Sam Speck (Emory University) and members of his laboratory for insightful discussions and technical advice, Minida Dowdy and Jennifer Perry for animal husbandry, Laura Bender at the cell imaging and microscopy core at The Winship Cancer Institute and Hong Yi at the Emory School of Medicine Microscopy Core. Supported by N.I.H (PHS grants R01 AI20211 and AI30363 to E.S.M) and (T32 HL069769 and F32 AI080175-01A1 to J.W.U.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Benedict CA. Viruses and the TNF-related cytokines, an evolving battle. Cytokine Growth Factor Rev. 2003;14:349–357. doi: 10.1016/s1359-6101(03)00030-3. [DOI] [PubMed] [Google Scholar]
- Benedict CA, Banks TA, Ware CF. Death and survival: viral regulation of TNF signaling pathways. Curr Opin Immunol. 2003;15:59–65. doi: 10.1016/s0952-7915(02)00018-3. [DOI] [PubMed] [Google Scholar]
- Brune W, Menard C, Heesemann J, Koszinowski UH. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science. 2001;291:303–305. doi: 10.1126/science.291.5502.303. [DOI] [PubMed] [Google Scholar]
- Bubic I, Wagner M, Krmpotic A, Saulig T, Kim S, Yokoyama WM, Jonjic S, Koszinowski UH. Gain of virulence caused by loss of a gene in murine cytomegalovirus. J Virol. 2004;78:7536–7544. doi: 10.1128/JVI.78.14.7536-7544.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bump NJ, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Li P, et al. Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- Cam M, Handke W, Picard-Maureau M, Brune W. Cytomegaloviruses inhibit Bak- and Bax-mediated apoptosis with two separate viral proteins. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.147. epublished ahead of print. [DOI] [PubMed] [Google Scholar]
- Ch’en IL, Beisner DR, Degterev A, Lynch C, Yuan J, Hoffmann A, Hedrick SM. Antigen-mediated T cell expansion regulated by parallel pathways of death. Proc Natl Acad Sci U S A. 2008;105:17463–17468. doi: 10.1073/pnas.0808043105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan FK, Shisler J, Bixby JG, Felices M, Zheng L, Appel M, Orenstein J, Moss B, Lenardo MJ. A role for tumor necrosis factor receptor-2 and receptor-interacting protein in programmed necrosis and antiviral responses. J Biol Chem. 2003;278:51613–51621. doi: 10.1074/jbc.M305633200. [DOI] [PubMed] [Google Scholar]
- Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, Chan FK. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofferson DE, Yuan J. Necroptosis as an alternative form of programmed cell death. Curr Opin Cell Biol. 2009 doi: 10.1016/j.ceb.2009.12.003. epublished ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicin-Sain L, Podlech J, Messerle M, Reddehase MJ, Koszinowski UH. Frequent coinfection of cells explains functional in vivo complementation between cytomegalovirus variants in the multiply infected host. J Virol. 2005;79:9492–9502. doi: 10.1128/JVI.79.15.9492-9502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicin-Sain L, Ruzsics Z, Podlech J, Bubic I, Menard C, Jonjic S, Reddehase MJ, Koszinowski UH. Dominant-negative FADD rescues the in vivo fitness of a cytomegalovirus lacking an antiapoptotic viral gene. J Virol. 2008;82:2056–2064. doi: 10.1128/JVI.01803-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clem RJ. The role of apoptosis in defense against baculovirus infection in insects. Curr Top Microbiol Immunol. 2005;289:113–129. doi: 10.1007/3-540-27320-4_5. [DOI] [PubMed] [Google Scholar]
- Clem RJ, Fechheimer M, Miller LK. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science. 1991;254:1388–1390. doi: 10.1126/science.1962198. [DOI] [PubMed] [Google Scholar]
- Crook NE, Clem RJ, Miller LK. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J Virol. 1993;67:2168–2174. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino DM, Bernardi P, Chieco-Bianchi L, Ciminale V. Mitochondria as functional targets of proteins coded by human tumor viruses. Adv Cancer Res. 2005;94:87–142. doi: 10.1016/S0065-230X(05)94003-7. [DOI] [PubMed] [Google Scholar]
- Datta S, Costantino N, Court DL. A set of recombineering plasmids for gram-negative bacteria. Gene. 2006;379:109–115. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- Declercq W, Vanden Berghe T, Vandenabeele P. RIP kinases at the crossroads of cell death and survival. Cell. 2009;138:229–232. doi: 10.1016/j.cell.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Degterev A, Hitomi J, Germscheid M, Ch’en IL, Korkina O, Teng X, Abbott D, Cuny GD, Yuan C, Wagner G, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N, Cuny GD, Mitchison TJ, Moskowitz MA, Yuan J. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- Degterev A, Yuan J. Expansion and evolution of cell death programmes. Nat Rev Mol Cell Biol. 2008;9:378–390. doi: 10.1038/nrm2393. [DOI] [PubMed] [Google Scholar]
- Feng S, Yang Y, Mei Y, Ma L, Zhu DE, Hoti N, Castanares M, Wu M. Cleavage of RIP3 inactivates its caspase-independent apoptosis pathway by removal of kinase domain. Cell Signal. 2007;19:2056–2067. doi: 10.1016/j.cellsig.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Festjens N, Vanden Berghe T, Cornelis S, Vandenabeele P. RIP1, a kinase on the crossroads of a cell’s decision to live or die. Cell Death Differ. 2007;14:400–410. doi: 10.1038/sj.cdd.4402085. [DOI] [PubMed] [Google Scholar]
- Festjens N, Vanden Berghe T, Vandenabeele P. Necrosis, a well-orchestrated form of cell demise: signalling cascades, important mediators and concomitant immune response. Biochim Biophys Acta. 2006;1757:1371–1387. doi: 10.1016/j.bbabio.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Friedman HM, Wang L, Fishman NO, Lambris JD, Eisenberg RJ, Cohen GH, Lubinski J. Immune evasion properties of herpes simplex virus type 1 glycoprotein gC. J Virol. 1996;70:4253–4260. doi: 10.1128/jvi.70.7.4253-4260.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Brenner C, Morselli E, Touat Z, Kroemer G. Viral control of mitochondrial apoptosis. PLoS Pathog. 2008;4:e1000018. doi: 10.1371/journal.ppat.1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Kroemer G. Necroptosis: a specialized pathway of programmed necrosis. Cell. 2008;135:1161–1163. doi: 10.1016/j.cell.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Goldmacher VS. Cell death suppression by cytomegaloviruses. Apoptosis. 2005;10:251–265. doi: 10.1007/s10495-005-0800-z. [DOI] [PubMed] [Google Scholar]
- Goldmacher VS, Bartle LM, Skaletskaya A, Dionne CA, Kedersha NL, Vater CA, Han JW, Lutz RJ, Watanabe S, Cahir McFarland ED, et al. A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc Natl Acad Sci U S A. 1999;96:12536–12541. doi: 10.1073/pnas.96.22.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Wang L, Miao L, Wang T, Du F, Zhao L, Wang X. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell. 2009;137:1100–1111. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135:1311–1323. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoebe K, Du X, Georgel P, Janssen E, Tabeta K, Kim SO, Goode J, Lin P, Mann N, Mudd S, et al. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- Holler N, Zaru R, Micheau O, Thome M, Attinger A, Valitutti S, Bodmer JL, Schneider P, Seed B, Tschopp J. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat Immunol. 2000;1:489–495. doi: 10.1038/82732. [DOI] [PubMed] [Google Scholar]
- Janssens S, Tinel A, Lippens S, Tschopp J. PIDD mediates NF-kappaB activation in response to DNA damage. Cell. 2005;123:1079–1092. doi: 10.1016/j.cell.2005.09.036. [DOI] [PubMed] [Google Scholar]
- Kaiser WJ, Offermann MK. Apoptosis induced by the toll-like receptor adaptor TRIF is dependent on its receptor interacting protein homotypic interaction motif. J Immunol. 2005;174:4942–4952. doi: 10.4049/jimmunol.174.8.4942. [DOI] [PubMed] [Google Scholar]
- Kaiser WJ, Upton JW, Mocarski ES. Receptor-interacting protein homotypic interaction motif-dependent control of NF-kappa B activation via the DNA-dependent activator of IFN regulatory factors. J Immunol. 2008;181:6427–6434. doi: 10.4049/jimmunol.181.9.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapadia SB, Levine B, Speck SH, Virgin HWt. Critical role of complement and viral evasion of complement in acute, persistent, and latent gamma-herpesvirus infection. Immunity. 2002;17:143–155. doi: 10.1016/s1074-7613(02)00369-2. [DOI] [PubMed] [Google Scholar]
- Kasof GM, Prosser JC, Liu D, Lorenzi MV, Gomes BC. The RIP-like kinase, RIP3, induces apoptosis and NF-kappaB nuclear translocation and localizes to mitochondria. FEBS Lett. 2000;473:285–291. doi: 10.1016/s0014-5793(00)01473-3. [DOI] [PubMed] [Google Scholar]
- Kattenhorn LM, Mills R, Wagner M, Lomsadze A, Makeev V, Borodovsky M, Ploegh HL, Kessler BM. Identification of proteins associated with murine cytomegalovirus virions. J Virol. 2004;78:11187–97. doi: 10.1128/JVI.78.20.11187-11197.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. TLR signaling. Semin Immunol. 2007;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Kelliher MA, Grimm S, Ishida Y, Kuo F, Stanger BZ, Leder P. The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity. 1998;8:297–303. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- Kepp O, Senovilla L, Galluzzi L, Panaretakis T, Tesniere A, Schlemmer F, Madeo F, Zitvogel L, Kroemer G. Viral subversion of immunogenic cell death. Cell Cycle. 2009;8:860–869. doi: 10.4161/cc.8.6.7939. [DOI] [PubMed] [Google Scholar]
- Kielczewska A, Pyzik M, Sun T, Krmpotic A, Lodoen MB, Munks MW, Babic M, Hill AB, Koszinowski UH, Jonjic S, et al. Ly49P recognition of cytomegalovirus-infected cells expressing H2-Dk and CMV-encoded m04 correlates with the NK cell antiviral response. J Exp Med. 2009;206:515–523. doi: 10.1084/jem.20080954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochem Biophys Res Commun. 2009;388:621–625. doi: 10.1016/j.bbrc.2009.08.062. [DOI] [PubMed] [Google Scholar]
- Leib DA, Machalek MA, Williams BR, Silverman RH, Virgin HW. Specific phenotypic restoration of an attenuated virus by knockout of a host resistance gene. Proc Natl Acad Sci U S A. 2000;97:6097–6101. doi: 10.1073/pnas.100415697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembo D, Donalisio M, Hofer A, Cornaglia M, Brune W, Koszinowski U, Thelander L, Landolfo S. The ribonucleotide reductase R1 homolog of murine cytomegalovirus is not a functional enzyme subunit but is required for pathogenesis. J Virol. 2004;78:4278–4288. doi: 10.1128/JVI.78.8.4278-4288.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubinski J, Wang L, Mastellos D, Sahu A, Lambris JD, Friedman HM. In vivo role of complement-interacting domains of herpes simplex virus type 1 glycoprotein gC. J Exp Med. 1999;190:1637–1646. doi: 10.1084/jem.190.11.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubinski JM, Wang L, Soulika AM, Burger R, Wetsel RA, Colten H, Cohen GH, Eisenberg RJ, Lambris JD, Friedman HM. Herpes simplex virus type 1 glycoprotein gC mediates immune evasion in vivo. J Virol. 1998;72:8257–8263. doi: 10.1128/jvi.72.10.8257-8263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack C, Sickmann A, Lembo D, Brune W. Inhibition of proinflammatory and innate immune signaling pathways by a cytomegalovirus RIP1-interacting protein. Proc Natl Acad Sci U S A. 2008;105:3094–3099. doi: 10.1073/pnas.0800168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzur M, Fleming P, Huang DC, Degli-Esposti MA, Andoniou CE. Virally mediated inhibition of Bax in leukocytes promotes dissemination of murine cytomegalovirus. Cell Death Differ. 2009;16:312–320. doi: 10.1038/cdd.2008.152. [DOI] [PubMed] [Google Scholar]
- McCartney SA, Colonna M. Viral sensors: diversity in pathogen recognition. Immunol Rev. 2009;227:87–94. doi: 10.1111/j.1600-065X.2008.00726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick AL. Control of apoptosis by human cytomegalovirus. Curr Top Microbiol Immunol. 2008;325:281–295. doi: 10.1007/978-3-540-77349-8_16. [DOI] [PubMed] [Google Scholar]
- McCormick AL, Meiering CD, Smith GB, Mocarski ES. Mitochondrial cell death suppressors carried by human and murine cytomegalovirus confer resistance to proteasome inhibitor-induced apoptosis. J Virol. 2005;79:12205–12217. doi: 10.1128/JVI.79.19.12205-12217.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick AL, Roback L, Mocarski ES. HtrA2/Omi terminates cytomegalovirus infection and is controlled by the viral mitochondrial inhibitor of apoptosis (vMIA) PLoS Pathog. 2008;4:e1000063. doi: 10.1371/journal.ppat.1000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard C, Wagner M, Ruzsics Z, Holak K, Brune W, Campbell AE, Koszinowski UH. Role of murine cytomegalovirus US22 gene family members in replication in macrophages. J Virol. 2003;77:5557–5570. doi: 10.1128/JVI.77.10.5557-5570.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Burns K, Hofmann K, Blancheteau V, Martinon F, Kelliher M, Tschopp J. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat Immunol. 2004;5:503–507. doi: 10.1038/ni1061. [DOI] [PubMed] [Google Scholar]
- Meylan E, Tschopp J. The RIP kinases: crucial integrators of cellular stress. Trends Biochem Sci. 2005;30:151–159. doi: 10.1016/j.tibs.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Michallet MC, Meylan E, Ermolaeva MA, Vazquez J, Rebsamen M, Curran J, Poeck H, Bscheider M, Hartmann G, Konig M, et al. TRADD Protein is an essential component of the RIG-like helicase antiviral pathway. Immunity. 2008;28:651–61. doi: 10.1016/j.immuni.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Miura M, Zhu H, Rotello R, Hartwieg EA, Yuan J. Induction of apoptosis in fibroblasts by IL-1 beta-converting enzyme, a mammalian homolog of the C. elegans cell death gene ced-3. Cell. 1993;75:653–660. doi: 10.1016/0092-8674(93)90486-a. [DOI] [PubMed] [Google Scholar]
- Newton K, Sun X, Dixit VM. Kinase RIP3 is dispensable for normal NF-kappa Bs, signaling by the B-cell and T-cell receptors, tumor necrosis factor receptor 1, and Toll-like receptors 2 and 4. Mol Cell Biol. 2004;24:1464–1469. doi: 10.1128/MCB.24.4.1464-1469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm NW, Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunol Rev. 2009;227:221–233. doi: 10.1111/j.1600-065X.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- Pazdernik NJ, Donner DB, Goebl MG, Harrington MA. Mouse receptor interacting protein 3 does not contain a caspase-recruiting or a death domain but induces apoptosis and activates NF-kappaB. Mol Cell Biol. 1999;19:6500–6508. doi: 10.1128/mcb.19.10.6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock JL, Virgin HWt. Latency, without persistence, of murine cytomegalovirus in the spleen and kidney. J Virol. 1995;69:1762–1768. doi: 10.1128/jvi.69.3.1762-1768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray CA, Black RA, Kronheim SR, Greenstreet TA, Sleath PR, Salvesen GS, Pickup DJ. Viral inhibition of inflammation: cowpox virus encodes an inhibitor of the interleukin-1 beta converting enzyme. Cell. 1992;69:597–604. doi: 10.1016/0092-8674(92)90223-y. [DOI] [PubMed] [Google Scholar]
- Rebsamen M, Heinz LX, Meylan E, Michallet MC, Schroder K, Hofmann K, Vazquez J, Benedict CA, Tschopp J. DAI/ZBP1 recruits RIP1 and RIP3 through RIP homotypic interaction motifs to activate NF-kappaB. EMBO Rep. 2009;10:916–922. doi: 10.1038/embor.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redwood AJ, Messerle M, Harvey NL, Hardy CM, Koszinowski UH, Lawson MA, Shellam GR. Use of a murine cytomegalovirus K181-derived bacterial artificial chromosome as a vaccine vector for immunocontraception. J Virol. 2005;79:2998–3008. doi: 10.1128/JVI.79.5.2998-3008.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy CR, Mocarski ES. Pathogen subversion of cell-intrinsic innate immunity. Nat Immunol. 2007;8:1179–1187. doi: 10.1038/ni1528. [DOI] [PubMed] [Google Scholar]
- Saederup N, Aguirre SA, Sparer TE, Bouley DM, Mocarski ES. Murine cytomegalovirus CC chemokine homolog MCK-2 (m131-129) is a determinant of dissemination that increases inflammation at initial sites of infection. J Virol. 2001;75:9966–9976. doi: 10.1128/JVI.75.20.9966-9976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saederup N, Lin YC, Dairaghi DJ, Schall TJ, Mocarski ES. Cytomegalovirus-encoded beta chemokine promotes monocyte-associated viremia in the host. Proc Natl Acad Sci U S A. 1999;96:10881–10886. doi: 10.1073/pnas.96.19.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Gale M., Jr Principles of intracellular viral recognition. Curr Opin Immunol. 2007;19:17–23. doi: 10.1016/j.coi.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Shultz LD, Lyons BL, Burzenski LM, Gott B, Chen X, Chaleff S, Kotb M, Gillies SD, King M, Mangada J, et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol. 2005;174:6477–6489. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
- Skaletskaya A, Bartle LM, Chittenden T, McCormick AL, Mocarski ES, Goldmacher VS. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc Natl Acad Sci U S A. 2001;98:7829–7834. doi: 10.1073/pnas.141108798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanger BZ, Leder P, Lee TH, Kim E, Seed B. RIP: a novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell. 1995;81:513–523. doi: 10.1016/0092-8674(95)90072-1. [DOI] [PubMed] [Google Scholar]
- Sun X, Lee J, Navas T, Baldwin DT, Stewart TA, Dixit VM. RIP3, a novel apoptosis-inducing kinase. J Biol Chem. 1999;274:16871–16875. doi: 10.1074/jbc.274.24.16871. [DOI] [PubMed] [Google Scholar]
- Sun X, Yin J, Starovasnik MA, Fairbrother WJ, Dixit VM. Identification of a novel homotypic interaction motif required for the phosphorylation of receptor-interacting protein (RIP) by RIP3. J Biol Chem. 2002;277:9505–9511. doi: 10.1074/jbc.M109488200. [DOI] [PubMed] [Google Scholar]
- Tabeta K, Georgel P, Janssen E, Du X, Hoebe K, Crozat K, Mudd S, Shamel L, Sovath S, Goode J, et al. Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc Natl Acad Sci U S A. 2004;101:3516–3521. doi: 10.1073/pnas.0400525101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandon R, Mocarski ES. Control of cytoplasmic maturation events by cytomegalovirus tegument protein pp150. J Virol. 2008;82:9433–9444. doi: 10.1128/JVI.00533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer JL, Schroter M, et al. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- Upton JW, Kaiser WJ, Mocarski ES. Cytomegalovirus M45 cell death suppression requires receptor-interacting protein (RIP) homotypic interaction motif (RHIM)-dependent interaction with RIP1. J Biol Chem. 2008;283:16966–16970. doi: 10.1074/jbc.C800051200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercammen D, Beyaert R, Denecker G, Goossens V, Van Loo G, Declercq W, Grooten J, Fiers W, Vandenabeele P. Inhibition of caspases increases the sensitivity of L929 cells to necrosis mediated by tumor necrosis factor. J Exp Med. 1998;187:1477–1485. doi: 10.1084/jem.187.9.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Gao X, Barrett JW, Shao Q, Bartee E, Mohamed MR, Rahman M, Werden S, Irvine T, Cao J, et al. RIG-I mediates the co-induction of tumor necrosis factor and type I interferon elicited by myxoma virus in primary human macrophages. PLoS Pathog. 2008;4:e1000099. doi: 10.1371/journal.ppat.1000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. Mechanisms of apoptosis regulation by viral oncogenes in infection and tumorigenesis. Cell Death Differ. 2006;13:1371–1377. doi: 10.1038/sj.cdd.4401941. [DOI] [PubMed] [Google Scholar]
- Wong WW, Gentle IE, Nachbur U, Anderton H, Vaux DL, Silke J. RIPK1 is not essential for TNFR1-induced activation of NF-kappaB. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.178. [DOI] [PubMed] [Google Scholar]
- Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- Yu PW, Huang BC, Shen M, Quast J, Chan E, Xu X, Nolan GP, Payan DG, Luo Y. Identification of RIP3, a RIP-like kinase that activates apoptosis and NFkappaB. Curr Biol. 1999;9:539–542. doi: 10.1016/s0960-9822(99)80239-5. [DOI] [PubMed] [Google Scholar]
- Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC, Dong MQ, Han J. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325:332–336. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.