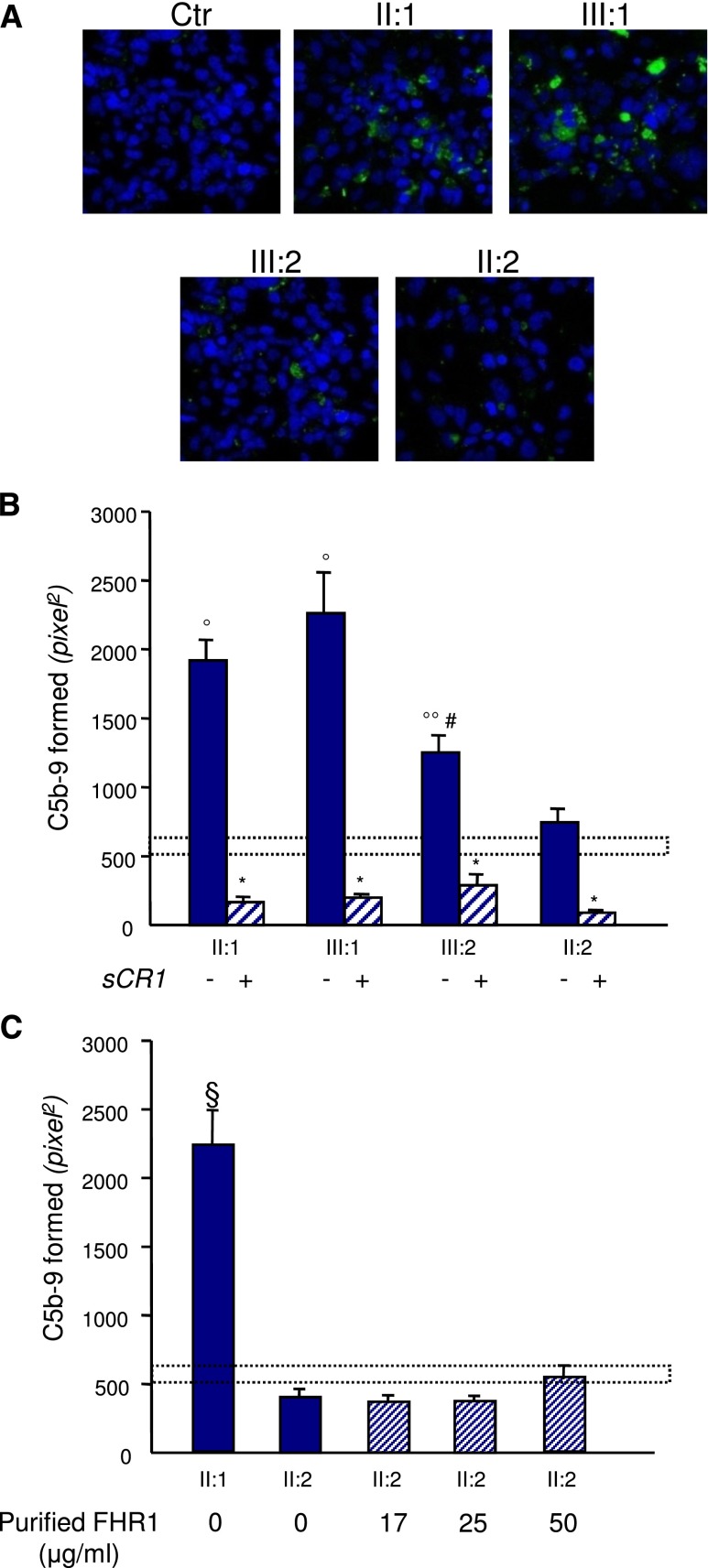
Figure 7.
Sera from CFHR1/CFH carriers cause C5b-9 deposition on endothelial cells. Panel A shows representative confocal microscopy images of C5b-9 staining (green) of ADP-activated HMEC-1 exposed for 4 hours to serum (diluted 1:2 in test medium) from a healthy control (Ctr), the proband (II:1; with the heterozygous duplication and the hybrid CFHR1/CFH gene), his affected daughter (III:1) and his unaffected son (III:2; both with the heterozygous duplication and the hybrid CFHR1/CFH gene), and his wife (II:2; with two normal copies of CFHR1). Original magnification, ×400. Blue indicates the 4′,6-diamidino-2-phenylindole staining of cell nuclei. (B) The graph shows the quantification of HMEC-1 area covered by C5b-9 deposits after incubation with serum from a healthy control, the proband, and relatives of the proband in the presence or not of the complement inhibitor sCR1 (150 µg/ml). The dotted rectangle shows the range of C5b-9 deposits induced by a pool of control sera. Data are means±SEMs. *P<0.001 versus serum untreated; °P<0.001, °°P<0.05 versus control; #P<0.05 versus patients. (C) Quantification of HMEC-1 area covered by C5b-9 deposits after incubation with serum from the proband or the healthy wife of the proband with or without the addition of increasing amounts of purified FHR1 from normal serum to mimic the product of one (17 and 25 μg/ml) or two (50 μg/ml) extra CFHR1 gene copies (details are in Supplemental Material). FHR1 addition to serum from the proband’s wife had no effect on C5b-9 deposits. Data are means±SEMs. The dotted rectangle shows the range of C5b-9 deposits induced by a pool of control sera. §P<0.001 versus all the others and versus controls.