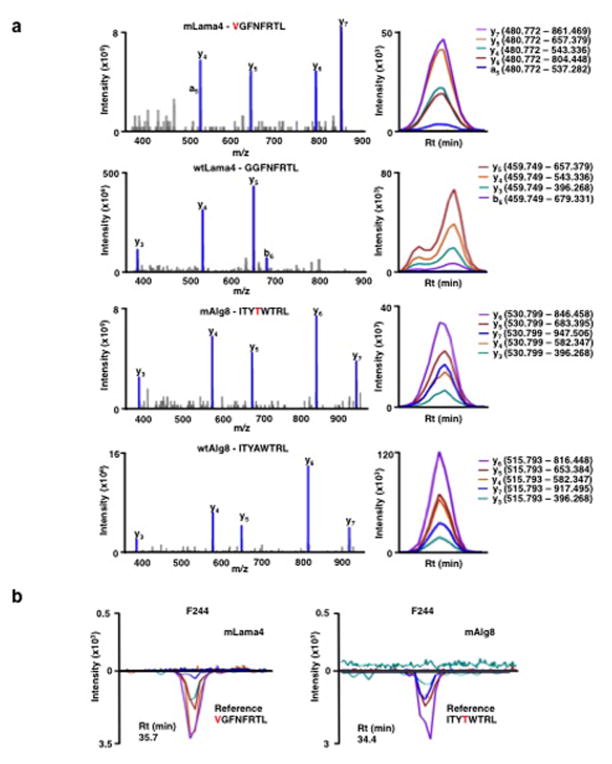
Extended Data Figure 6. Generation of SRM assay library for the detection of mutant H-2Kb peptides on d42m1-T3.
a, SRM transitions were optimized for 51 of the 62 top predicted H-2Kb peptides. The 51 peptides chosen were selected based on having physiochemical properties that would allow their detection by MS if present. Only Lama4 and Alg8 are shown here for simplicity. The 51 peptides were synthesized and LC-MS/MS acquisition was performed on each peptide to determine the best collision energy and to obtain the full fragment ion spectrum (left panel); three to seven of the highest intensity peaks were selected to be built into SRM transitions. Optimal SRM transitions displayed as extracted ion chromatograms are shown (right panel). Q1 – Q3 transitions are indicated in parenthesis. The mutated amino acid in the peptide sequence is marked in red. b, F244 tumour cells, which lack the mLama4 and mAlg8 d42m1-T3 epitope, lack detectable mLama4 or mAlg8 in complex with H-2Kb as assessed by SRM.