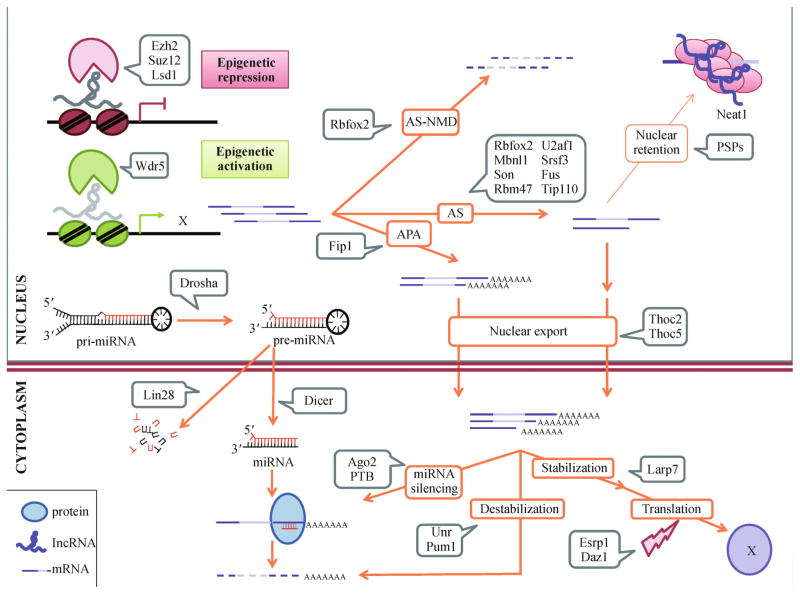
Figure 1.
RBPs with important roles in ESC maintenance and/or differentiation are depicted according to the RNA metabolism step in which they are implicated. Ezh2, Suz12 and Lsd1 repress transcription whereas Wdr5 activates it, binding to chromatin through lncRNAs. Rbfox2 is implicated both in AS and AS-NMD, whereas Mbnl1, Son, Rbm47, U2af1, Srsf3 and Fus have only been reported to regulate AS. Fip1 mediates APA. Once processed, transcripts can either be retained in paraspeckles by PSPs or exported to the cytoplasm. Thoc2 and Thoc5 regulate the transport of important pluripotency-related transcripts in ESC. In the cytoplasm, transcripts are stabilized and transcribed to proteins or targeted for destabilization and/or miRNA silencing by Unr and Pum1 or Ago2 and PTB, respectively. Translation efficiency can be also subjected to regulation, through proteins such as Larp7 (stabilizer) or Esrp1 and Dazl (reduce ribosomal loading). Processing of pri-miRNAs by Drosha and pre-miRNA by Dicer gives rise to mature miRNAs available in the cytoplasm for transcript silencing. Lin28 affects its targets through targeting pre-miRNAs for degradation, therefore reducing the steady-state levels of miRNAs. From top to bottom: AS-NMD: alternative splicing-coupled nonsense-mediated decay, AS: alternative splicing; APA: alternative polyadenylation; PSPs: paraspeckle proteins; pri-miRNA: primary microRNA transcript; pre-miRNA: precursor microRNA transcript; miRNA: microRNA.