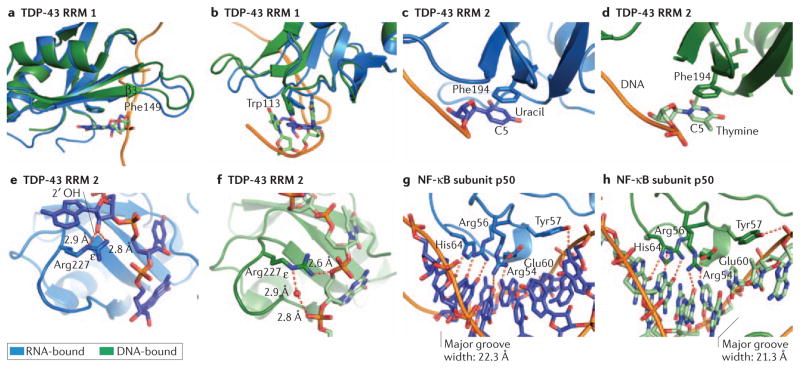
Figure 4. The structural basis for dual DNA and RNA recognition by TDP-43 and by the NF-κB subunit p50.
Protein-RNA structures are shown in blue and protein-DNA structures in green, with protein in the darker shade. π-stacking interactions play a prominent role in both the ssDNA and ssRNA binding activities of TDP-43: a–d. Phe149 (a) and Trp113 (b) within the first RRM of TDP43 stack with both RNA and DNA bases. c, d. In the second RRM of TDP-43, Phe194 is capable of recognizing both uracil in RNA and thymine in DNA; the additional methyl group at C5 in thymine does not contribute to nucleic acid specificity. e. When bound to RNA, both the terminal amine and ε nitrogen of Arg227 in the second RRM of TDP-43 contact a 2′OH on the RNA backbone. f. In contrast, these same groups can also make contacts with the DNA backbone, both directly and through water mediated hydrogen bonding. g, h. The p50 subunit of NF-κB makes strikingly similar base-specific contacts when bound to double-stranded DNA (g) or an RNA aptamer (h). This is due in large part to the similar secondary structure and chemical moieties presented by the RNA and DNA. Major groove width was calculated by 3DNA using phosphate-phosphate distances92.