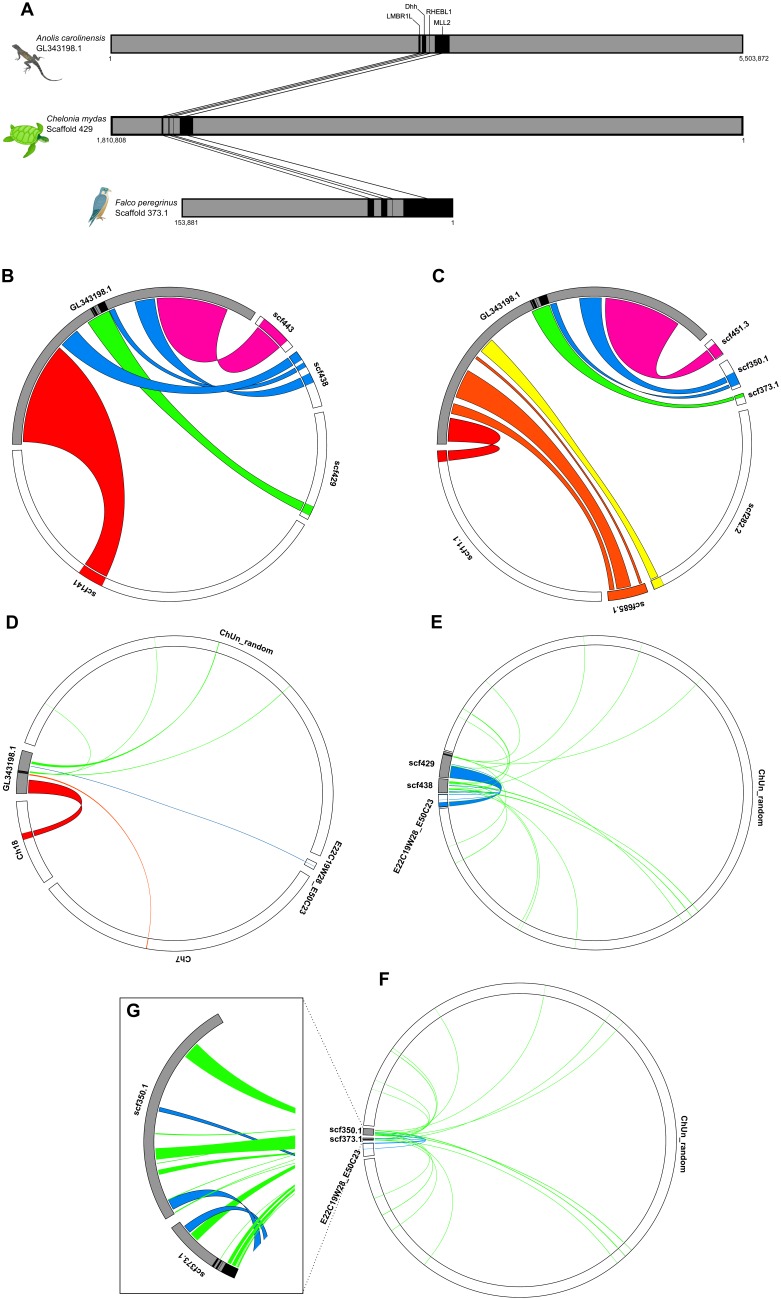
Figure 3. Homology between the Anolis carolinensis Dhh gene synteny and the Chelonia mydas, Falcon peregrinus and Gallus gallus genomes, using nucleotide information.
(A) The tetrapod LMBR1L–Dhh-RHEBL1-MLL2 gene cluster is found on the scaffold GL343198.1 scaffold of the A. carolinensis assembly (anoCar 2.0 [105]), and a similar cluster is also found on the 373.1 scaffold of the F. peregrinus genome assembly. (B) The four main C. mydas scaffolds show great homology for specific regions of the lizard GL343198.1 scaffold, (C) six on the F. peregrinus genome, and (d) on G. Gallus genome homology is found on two macrochromosomes, a linkage group and the Un_random chromosome. (e) The 429 and 439 C. mydas scaffold have high homology with a great part of the chicken E22C19W28_E50C23 linkage group and several random regions of the chicken Un_random chromosome. (F) The 350.1 and 373.1 F. peregrinusscaffold have high homology with several random regions of the G. gallus Un_random chromosome. (E) Hits for the F. peregrinus cluster are found on G. gallus genome mainly for the MLL2 gene.