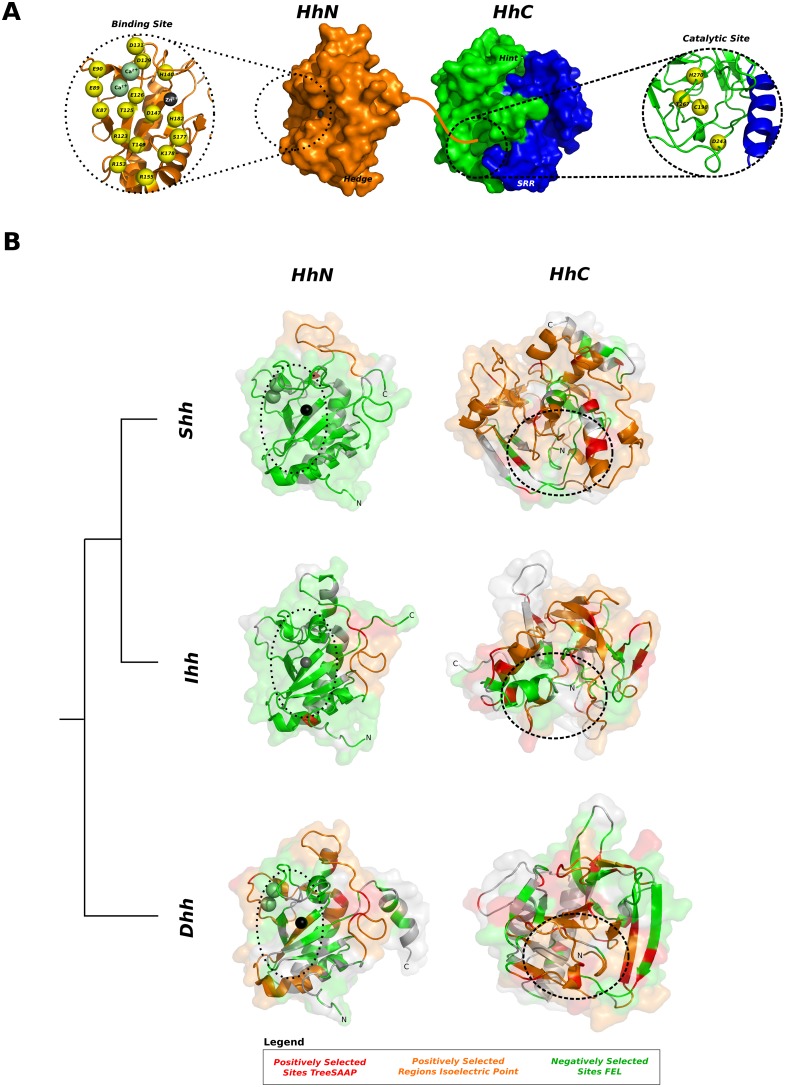
Figure 9. Tridimensional arrangement of negatively and positively regions in the Hedge domain of vertebrate Hedgehog proteins.
(A) Tridimensional representation of Hedge (PDB: 3HO5) and Hog (Shh modelled by homology) domains, coloured according to key identified regions. A straight orange line denotes how both domains may be linked in the pro-protein. Key residues important for binding and forming the catalytic site are represented in yellow spheres, numbered according to the human Shh protein [72], [113], [114]. (B) Tridimensional arrangement of negatively and positively selected regions across the Hedge (HhN) and Hog (HhC) domains. Proteins (ShhN: PDB 3HO5, DhhN: PDB 2WFR; IhhN: PDB 3K7G) represented in grey cartoon with transparent surface. Calcium ions are not represented on the IhhN peptide due to the absence of the ions on the PDB file. Negatively selected sites (green) identified with FEL, positively selected regions for the amino acid isoelectric point property (orange) and positively selected sites (red) identified with TreeSAAP are shown for each paralog domain. A dashed circle denotes the position of the zinc/calcium binding site and the catalytic site.