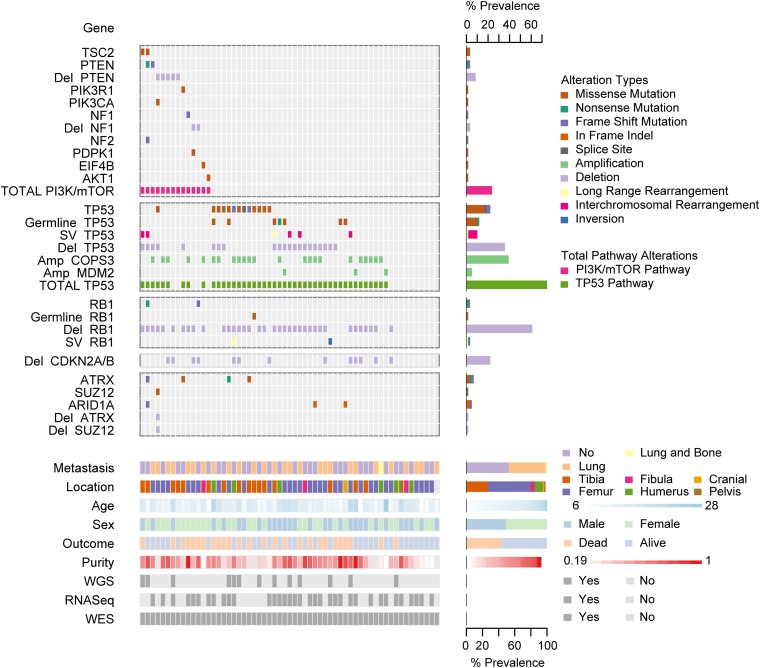
Fig. 1.
Summary of sequencing results highlighting alterations in the PI3K/mTOR pathway, TP53, RB1, and TP53 and RB1 interacting genes, demographic and clinical variables, and sequencing characteristics. Each column represents a patient sample. The bottom section of the graph indicates the sequencing characteristics, demographic, and clinical data for each patient. The top section of the graph indicates the types of alteration for each gene or pathway per sample. Copy number alterations for PI3K/mTOR pathway genes, ATRX, SUZ12, and ARID1A were determined with the heuristic algorithm PHIAL. Copy number alterations for TP53, RB1, and TP53 and RB1 interacting genes were determined with GISTIC2. Amp, amplification; Del, deletion; SV, structural variation; long-range rearrangement, intrachromosomal rearrangements >1 Mb.