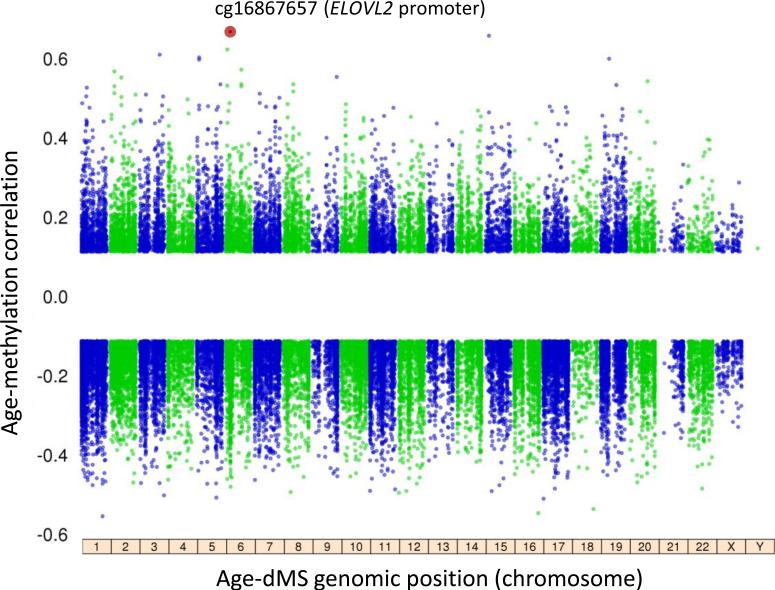
Figure 1. The aging methylome in 1,264 monocyte samples.
The analysis of age and methylation in 1,264 CD14+ monocyte samples included 448,523 CpG sites, of which 37,911 had methylation associated with age (age-dMS; 26,159 negatively associated, 11,752 positively associated, FDR<0.001). The most significant age-dMS (red circle, cg16867657, prho = 0.66, FDR = 3.65×10−140) was detected on chromosome 6 in the ELOVL2 (ELOVL fatty acid elongase 2) promoter. The partial correlation between CpG methylation and age is shown on y-axis, compared to CpG genomic location (by chromosome, x-axis). Linear regression analysis also included the following covariates: race, sex, site of data collection, microarray chip, and residual sample contamination with non-targeted cells (see Methods).