A unique mitochondrial ion channel affects organelle physiology and its lack is associated with senescence in the model plant Arabidopsis.
Abstract
Since the discovery of 20 genes encoding for putative ionotropic glutamate receptors in the Arabidopsis (Arabidopsis thaliana) genome, there has been considerable interest in uncovering their physiological functions. For many of these receptors, neither their channel formation and/or physiological roles nor their localization within the plant cells is known. Here, we provide, to our knowledge, new information about in vivo protein localization and give insight into the biological roles of the so-far uncharacterized Arabidopsis GLUTAMATE RECEPTOR3.5 (AtGLR3.5), a member of subfamily 3 of plant glutamate receptors. Using the pGREAT vector designed for the expression of fusion proteins in plants, we show that a splicing variant of AtGLR3.5 targets the inner mitochondrial membrane, while the other variant localizes to chloroplasts. Mitochondria of knockout or silenced plants showed a strikingly altered ultrastructure, lack of cristae, and swelling. Furthermore, using a genetically encoded mitochondria-targeted calcium probe, we measured a slightly reduced mitochondrial calcium uptake capacity in the knockout mutant. These observations indicate a functional expression of AtGLR3.5 in this organelle. Furthermore, AtGLR3.5-less mutant plants undergo anticipated senescence. Our data thus represent, to our knowledge, the first evidence of splicing-regulated organellar targeting of a plant ion channel and identify the first cation channel in plant mitochondria from a molecular point of view.
In vertebrates, ionotropic glutamate receptors (iGluRs in animals) are ligand-gated cation channels that mediate the majority of the excitatory neurotransmission in the central nervous system (Dingledine et al., 1999). In the model plant Arabidopsis (Arabidopsis thaliana), 20 genes encoding homologs of animal iGluRs have been identified (Lam et al., 1998). According to phylogenetic analyses, the Arabidopsis GLUTAMATE RECEPTOR (AtGLR) homologs can be subdivided into three separate subgroups (Chiu et al., 1999, 2002). Some evidence for the channel-forming ability by plant ionotropic glutamate receptors (iGLRs) has been obtained only recently, and only for AtGLR3.4 and AtGLR1.4 expressed in heterologous systems (Vincill et al., 2012; Tapken et al., 2013). Studies with transgenic plants suggested roles of members of the plant GLR family in Ca2+ fluxes (AtGLR2; Kim et al., 2001), coordination of mitotic activity in the root apical meristem (Li et al., 2006), regulation of abscisic acid biosynthesis and water balance (AtGLR1.1; Kang and Turano, 2003; Kang et al., 2004), carbon/nitrogen sensing (AtGLR1.1; Kang and Turano, 2003), resistance against fungal infection (Kang et al., 2006), leaf-to-leaf wound signaling (Mousavi et al., 2013), and lateral root initiation (Vincill et al., 2013). Application of antagonists and agonists of animal iGluRs revealed that plant GLRs might be involved in the regulation of root growth and branching (Walch-Liu et al., 2006), in light signal transduction (Lam et al., 1998), and in the response to aluminum (Sivaguru et al., 2003). In various plant cell types, the agonists Glu- and Gly-induced plasma membrane depolarization and a rise in intracellular Ca2+ concentration that were inhibited by blockers of nonselective cation channels (NSCCs) and by antagonists of animal iGluRs (Dennison and Spalding, 2000; Dubos et al., 2003; Meyerhoff et al., 2005; Krol et al., 2007; Kwaaitaal et al., 2011; Michard et al., 2011). Furthermore, Glu-activated cation currents in patch-clamped root protoplasts were inhibited by NSCC blockers such as La3+ and Gd3+ (Demidchik et al., 2004). Therefore, it was proposed that plant iGLRs can form Ca2+-permeable NSCCs, are inhibited by animal iGluR antagonists, and might contribute to the shaping of plant Ca2+ signaling (McAinsh and Pittman, 2009). Studies using AtGLR3.3 mutant plants showed that intracellular Ca2+ rise and membrane depolarization induced by Glu in Arabidopsis hypocotyls and root cells are correlated with the presence of AtGLR3.3 (Qi et al., 2006; Stephens et al., 2008).
However, most plant iGLRs, when expressed in heterologous systems, do not give rise to any current (e.g. in Xenopus spp. oocytes) or are toxic to host cells (e.g. in mammalian cells; Davenport, 2002). Recently, to examine whether AtGLR homologs possess functional ion channel domains, Tapken and Hollmann (2008) transplanted the pore loop together with two adjacent intracellular loops of 17 AtGLR subunits into two rat iGluR subunits and tested the resulting chimeric receptors for ion channel activity in the heterologous expression system Xenopus spp. oocyte. They showed that AtGLR1.1 and AtGLR1.4 have functional ion pore domains. The AtGLR1.1 pores are permeable to Na+, K+, and Ca2+ and are blocked by the nonspecific cation channel blocker La3+ (Tapken and Hollmann, 2008). Recent work has demonstrated that the expression of full-length AtGLR1.4 in oocytes gives rise to an amino acid-activated, nonselective, calcium-permeable channel that was found to be inhibited by the animal iGluR modulators 6,7-dinitroquinoxaline-2,3-dione and 6-cyano-7-nitroquinoxaline-2,3-dione (Tapken et al., 2013).
The study of these channels has so far been restricted to those members that are located in the plasma membrane and were proved to be functional in the expression systems used. Instead, various localization prediction tools suggest that some of the plant GLRs might have chloroplast and mitochondrial targeting. In general, determining the subcellular localization of a protein is an important step toward understanding its function. We recently reported the localization of GLR3.4 to the inner chloroplast membrane (Teardo et al., 2011), which was also shown to harbor a 6,7-dinitroquinoxaline-2,3-dione-sensitive, calcium-permeable channel activity (Teardo et al., 2010). No other studies have addressed the eventual subcellular localization of other putative Glu receptors.
In this work, we show that an isoform of GLR3.5 is efficiently targeted to the mitochondria. Functional expression of the channel in this organelle is indicated by the fact that its absence in knockout plants leads to a dramatically altered ultrastructure of mitochondria that impacts the plant physiology, ultimately leading to an anticipated senescence.
RESULTS
Cloning of the Two Splicing Variants of AtGLR3.5
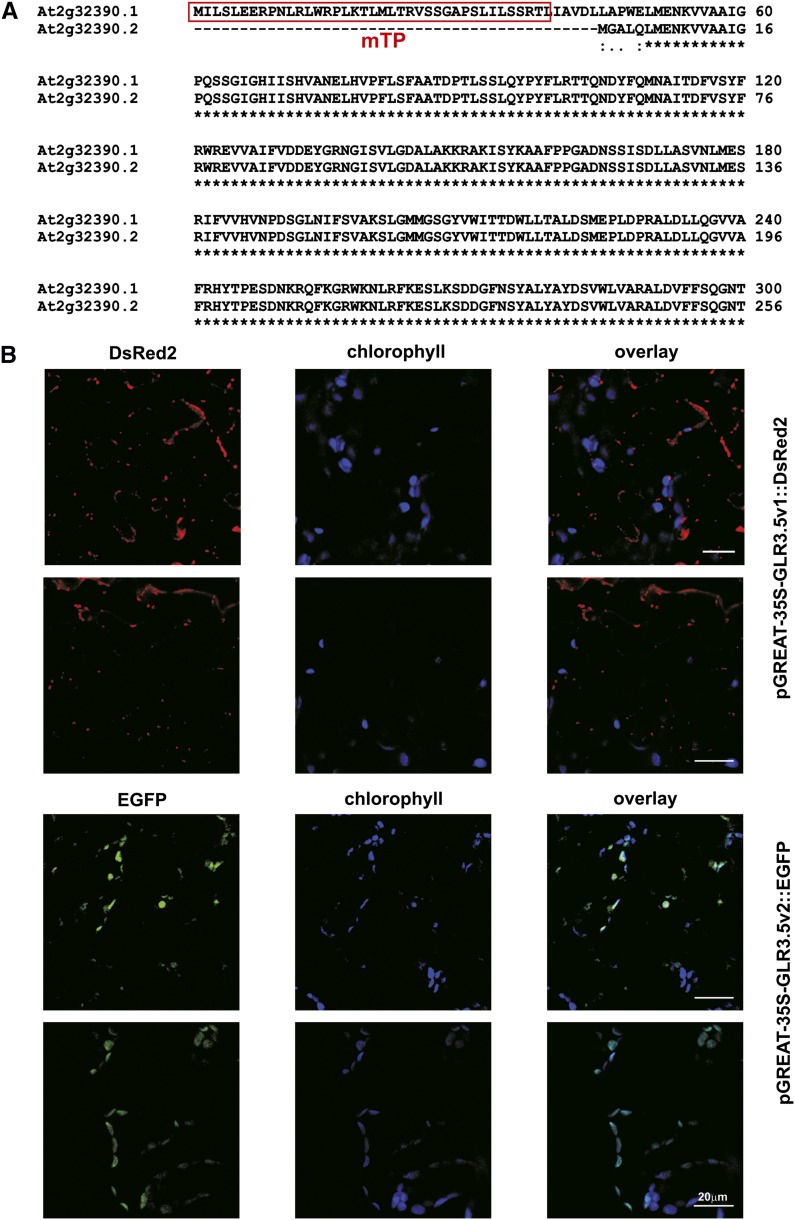
The Arabidopsis Glu receptor AtGLR3.5 is encoded by the At2g32390 gene that is transcribed in two splicing variants, NM_128798 (isoform 1) and NM_001036387 (isoform 2), corresponding to the gene models At2g32390.1 and At2g32390.2, respectively. The 5′ sequence is affected by the splicing, with the consequent modification of the putative targeting peptide between long (isoform 1) and short (isoform 2) translated proteins (Fig. 1A). Although a third gene model has been generated in The Arabidopsis Information Resource, only two isoforms have been demonstrated to be expressed so far. Their mRNA sequences correspond to accession numbers AF170494 and AY495449.
Figure 1.
Splicing variants of AtGLR3.5. A, Alignment of N-terminal regions of the two AtGLR3.5 isoforms. Amino acid sequences are shown. The predicted mitochondrial targeting sequence (TargetP) is in the red box. According to ChloroP1.1, the chloroplast-targeting sequence is 82 amino acids long. B, Subcellular localization of the AtGLR3.5 isoforms with predicted mitochondrial (isoform 1) and chloroplast (isoform 2) targeting. Expression of pGREAT-2x35S-GLR3.5v1::DsRed2 (top) and pGREAT-2x35S-GLR3.5v2::EGFP (bottom) in 4-week-old Arabidopsis leaves is shown. The isoform1-DsRed2 fusion protein is located in highly motile structures resembling mitochondria (Supplemental Movies S1 and S2). Results shown are representative of three independent experiments giving the same results. Bars = 20 µm.
At the protein level, isoform 1 (NP_565743.1) shows a putative signal peptide for the localization to the mitochondria that is missing in isoform 2 (NP_001031464.1; Aramemnon database [http://aramemnon.uni-koeln.de] and ChloroP [Emanuelsson et al., 2007]). To confirm the predicted localization of the two isoforms to the respective organelles, we isolated and cloned the complementary DNAs (cDNAs) corresponding to transcripts NM_128798 and NM_001036387 from leaf RNA by reverse transcription (RT)-PCR using the primers listed in Supplemental Table S1. As the sequence recognized by the primers, corresponding to the beginning of the coding sequence of isoform 1, is also present at the 5′ untranslated region of the isoform 2 transcript, the PCR product comprised both cDNAs. We designed a primer spanning the nine nucleotides in positions 150 to 158 of isoform 2 that are missing in isoform 1 to discriminate between the two isoforms. Thus, the Escherichia coli clones harboring the two different isoforms have been separated by PCR.
AtGLR3.5 Isoform 1 Is Located in the Mitochondria and Isoform 2 Targets Chloroplasts
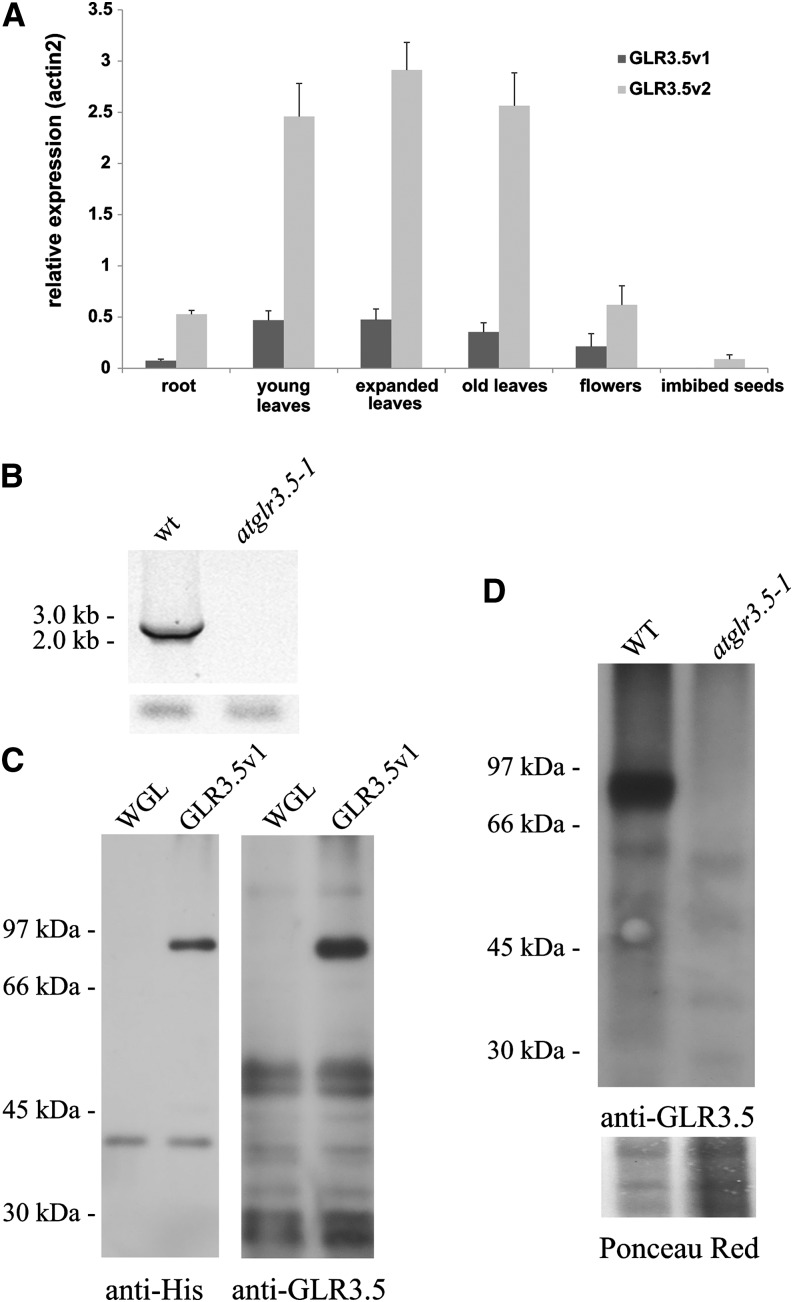
The coding sequences of the two isoforms have been cloned into binary vectors developed in our laboratory (Carraretto et al., 2013; pGREAT-2x35S-EGFP and pGREAT-2x35S-DsRed2) and transformed into Agrobacterium tumefaciens strain GV3101 for subsequent Arabidopsis leaf agroinfiltration. Figure 1B shows the targeting of isoform 1 (GLR3.5v1) to highly motile structures in the cytoplasm resembling mitochondria visualized using mitochondria-targeted mCherry (Nelson et al., 2007), both for size and motility (Fig. 1B; Supplemental Movies S1 and S2). The fluorescence of the GFP-fused short isoform (GLR3.5v2) instead colocalizes with the autofluorescence of chlorophyll in the chloroplasts (with a Pearson coefficient greater than 0.8 at the chloroplast level; Fig. 1B). The relative expression of the mitochondrial isoform over the plastidial one was assessed in several tissues (Fig. 2A), showing an overall lower expression level for the mitochondrial GLR3.5.
Figure 2.
Mitochondrial localization of splicing variant 1. A, Relative expression of the two transcript variants. Analysis of the relative amount of each variant in different tissues/organs of 4-week old Arabidopsis wild-type (WT) Col-0 plants is shown. ACTIN2 (At3g18780) was used as a reference gene. B, Transcript analysis of the Arabidopsis SALK mutant atglr3.5-1. PCR was performed with the primers reported in Supplemental Table S1. A positive control of transcript content (ACTIN) is shown (bottom). This result was also confirmed on another set of plants. C, AtGLR3.5 isoform 1 was expressed in vitro using a wheat germ lysate kit. Empty reaction mix (WGL) and mix following expression of the protein (G35V1) were loaded on SDS-PAGE gels and assayed with anti-His tag and anti-GLR3.5 antibodies. D, Western blot of mitochondria purified from wild-type and knockout plants (atglr3.5-1; 100 µg each) assayed with anti-GLR3.5 antibody. The membrane shown (bottom) was colored with Ponceau Red and shows comparable loading. Results in C and D are representative of three experiments.
Knocking Out and Silencing of the At2g32390 Gene Alters the Mitochondria Ultrastructure and Dimension
Two knockout homozygous mutant lines have been obtained from the Nottingham Arabidopsis Stock Centre: N656359 (SALK_035264C, atglr3.5-1) and N661846 (SALK_023880C, atglr3.5-2). Both lines have been tested for kanamycin resistance, transfer DNA insertion, and the absence of At2g3290 gene expression. Using different primer combinations to discriminate between the two isoforms, we observed that only the atglr3.5-1 mutant lacks the transcripts (Fig. 2B).
Given that isoform 1 is the first GLR located in the mitochondria, we focused our attention on elucidating the physiological role of AtGLR3.5 in this organelle. First, we selected a homozygous knockout line (N656359, SALK_035264C, atglr3.5-1) as the unique line not expressing the gene (Fig. 2B). We then checked the presence or absence of the protein in the mitochondria isolated from wild-type or knockout plants using an antibody, which is able to specifically recognize the recombinant AtGLR3.5 (Fig. 2C) with an apparent molecular mass of approximately 90 kD, which is close to the predicted apparent molecular mass. We clearly detected an approximately 88-kD band in the isolated mitochondria (enriched in cytochrome c; Supplemental Fig. S1) from wild-type Columbia-0 (Col-0) plants, while the protein, as expected, was completely missing in the mutant mitochondria (Fig. 2D).
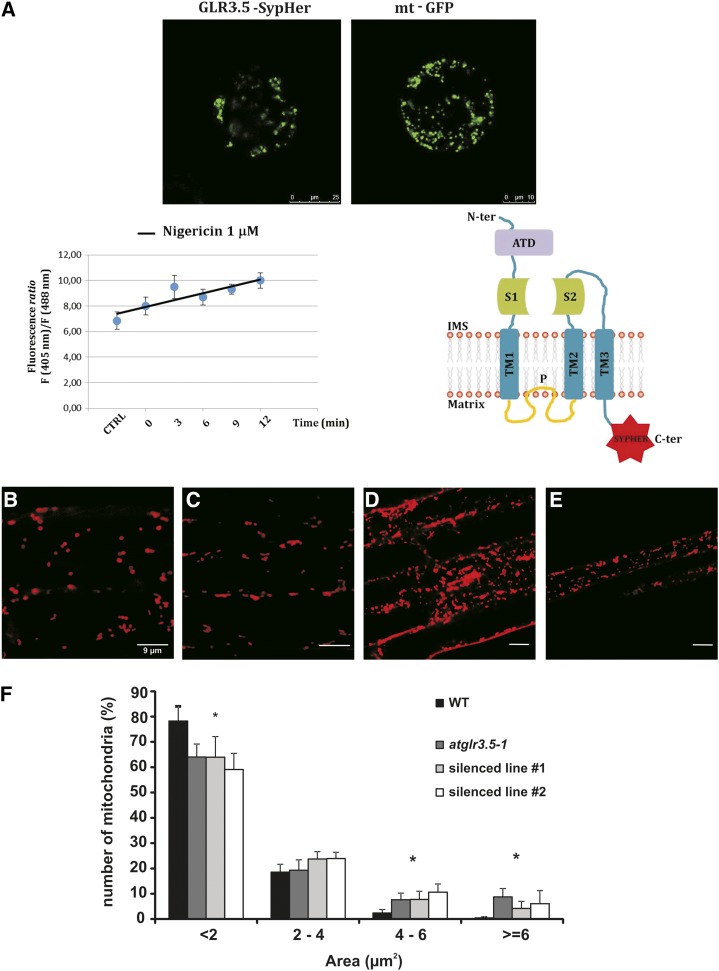
In order to assess the localization of GLR3.5 within the mitochondria and its membrane topology, we used GLR3.5v1 in fusion with a pH-sensitive ratiometric probe, SypHer. The mitochondria-targeted version of this probe has been reported to monitor dynamic changes in the mitochondrial pH gradient in HeLa cells (Poburko et al., 2011). In mitochondria, the proton motive force comprises an electrical component and a chemical component, the transmembrane pH gradient (approximately 0.8, alkaline inside). Thus, the observation that SypHer fused to the C terminus of GLR3.5v1 was able to sense matrix acidification induced by H+ ionophores (Fig. 3A) indicated that the C-terminal part of the protein is facing the matrix, meaning also that GLR3.5 is inserted into the inner mitochondrial membrane.
Figure 3.
Topology of GLR3.5 and differences in mitochondrial dimensions in the absence of GLR3.5. A, Confocal images of protoplasts expressing GLR3.5-SypHer (left) and mitochondrially targeted GFP (right). The effect of acidification on SypHer fluorescence ratio (via the addition of nigericin to the cells) is shown (bottom left; n = 4 ± se). Nigericin causes immediate acidification. Predicted topology (ATD, amino-terminal domain; S1 and S2, Glu-binding domains) is shown (bottom right). B to E, TMRM staining of intact 10-d-old seedling roots from wild-type (WT), knockout (atglr3.5-1), and silenced plants (#1 and #2). F, Statistical analysis of the distribution of mitochondria size revealing significant differences (**P < 0.01, *P < 0.05, Student’s t test) between the two genotypes.
Next, the number, dimension, and motility of mitochondria were analyzed using the cationic membrane-potential sensitive dye tetramethylrhodamine methyl ester (TMRM) in roots of 10-d-old plants. No significant differences were seen in either the number or motility of these organelles when comparing wild-type (Fig. 3B; Supplemental Movie S3) and knockout (Fig. 3C; Supplemental Movie S4) or silenced (Fig. 3, D and E) plants. Posttranscriptional silencing was performed as described by Carraretto et al. (2013; Supplemental Fig. S2). Moreover, the clear TMRM detection in wild-type and knockout or silenced plants suggests that, in both cases, the mitochondria in these young plants were sufficiently energized to take up the potentiometric dye. Interestingly, in all lines lacking GLR3.5, we observed a significantly higher percentage of mitochondria with an increased size, which was found by measuring the area of each single organelle. In particular, mutant plants displayed a higher percentage of organelles with a size larger than 4 μm2 (Fig. 3F).
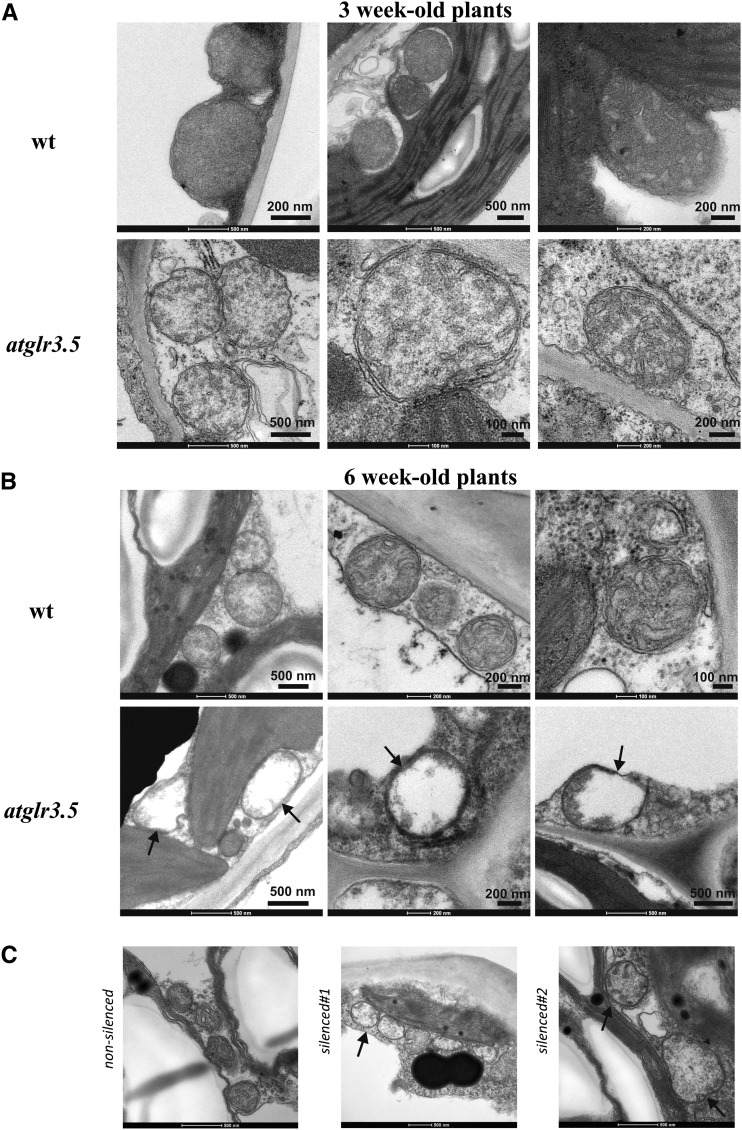
When we investigated the ultrastructure of these lines at the cellular level by transmission electron microscopy (TEM), we observed that in 3-week-old knockout plants, part of the mitochondria displayed enlargement and partial loss of cristae (Fig. 4A). This result is in agreement with the larger size of part of the mitochondria observed with TMRM in 10-d-old root cells. At 6 weeks, this phenotype became even more pronounced: most mitochondria were less electron dense and swollen, and lacked cristae (Fig. 4B; Supplemental Fig. S3). Altered mitochondrial ultrastructure was evident also in the silenced plants (Fig. 4C; Supplemental Fig. S3). In contrast to mitochondria, no dramatic alterations were noticeable with respect to the wild type for chloroplasts, where isoform 2 is located (Supplemental Fig. S3), possibly due to the presence in these organelles of other iGLRs such as AtGLR3.4 (Teardo et al., 2010, 2011), which might compensate for the loss of function of AtGLR3.5.
Figure 4.
Mitochondria from atglr3.5-1 and silenced plants show altered ultrastructure. A and B, TEM scans of mitochondria from 3-week-old (A) and 6-week-old (B) wild-type (wt) Col-0 and knockout plants. Representative images from wild-type (top rows) and knockout plants (atglr3.5; bottom rows) are shown. C, Mitochondria from nonsilenced and two independent silenced lines.
AtGLR3.5 Mutants Show Accelerated Senescence
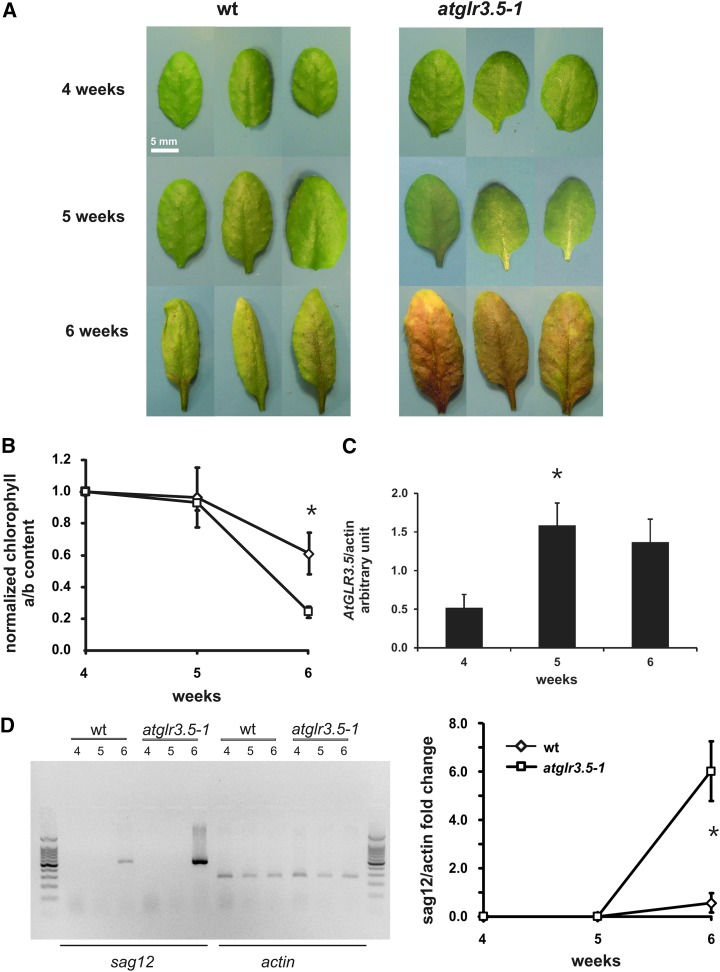
An altered mitochondrial size/structure is one of the characteristics of plants subjected to stress and may lead to cell death and senescence (Gan, 2007; Scott and Logan, 2008). Therefore, we investigated whether this aspect is affected in knockout plants. Mutant plants grow similarly to the wild-type plants up to the bolting stage, undergoing accelerated senescence in the subsequent phases. In fact, 5 to 6 weeks after germination, the knockout mutants show a more pronounced decrease in chlorophyll content in older leaves compared with the wild-type plants (Fig. 5, A and B). Interestingly, we also observed an increase in the expression of the AtGLR3.5 gene in 5-week-old wild-type plants (Fig. 5C). To prove that the observed changes were associated with senescence, the expression level of the senescence-associated gene SAG12, a well-known senescence marker, was assessed. In 6-week-old plants, the expression level of SAG12 was remarkably higher in atglr3.5-1 mutants than in the wild-type plants (Fig. 5D). Both these results strongly suggest an earlier onset of senescence in knockout plants.
Figure 5.
AtGLR3.5 knockout mutant plants show an accelerated senescence. A, Representative leaves from wild-type (wt) and knockout (atglr3.5-1) plants at 4, 5, and 6 weeks. B, The chlorophyll content in mutant leaves (n = 5) decreased faster than that of wild-type leaves 5 weeks after sowing. C, Increase in AtGLR3.5 gene expression during development. The results of three independent transcript analyses (±sd) obtained using densitometry are shown. D, Significant difference in SAG12 expression can be observed in the same plants (n = 3). The same results as shown here were obtained in another set of independent experiments. Left, representative gel of transcript analysis regarding SAG12 induction in wild-type and atglr3.5-1 plants at 4, 5, and 6 weeks. ACTIN transcript analysis was used as an internal control. A 100-bp DNA ladder was loaded. Right, graph showing statistically significant differences (asterisks) in SAG12 expression.
Mitochondrial Calcium Uptake in atglr3.5-1 Mutants
Given that some members of the family 3 iGLRs have been proven to allow calcium flux and that mitochondria are known to behave as calcium sinks in eukaryotic cells (De Stefani et al., 2011; Loro et al., 2012; Rizzuto et al., 2012), our next purpose was to gain evidence that calcium fluxes take place across AtGLR3.5. Our attempts to perform patch-clamp experiments directly on mitochondrial membranes (mitoplasts [i.e. swollen mitochondria]) of Arabidopsis, however, failed because of the very small size of these mitoplasts when compared with mammalian ones (with a diameter of 2–3 µm), where this technique, although demanding, is applicable. Next, we investigated the channel activity of the cell-free expressed AtGLR3.5 protein (Fig. 2C) following its incorporation to the planar lipid bilayer according to the procedure already successfully applied in the case of the mammalian mitochondrial calcium uniporter (De Stefani et al., 2011). The fact that no channel activity could be observed in this in vitro system (data not shown) clearly suggests either the need of an accessory protein to ensure correct folding or the need of an auxiliary, unknown partner for the reconstitution of the channel activity in vitro.
Therefore, we directly compared the mitochondrial calcium uptake kinetics in root seedlings from 7-d-old wild-type and mutant plants. Given the large negative membrane potential on the matrix side, we expected calcium influx into the mitochondria through GLR3.5. We crossed wild-type and atglr3.5-1 plants with Arabidopsis Col-0 plants harboring the mitochondria-targeted, genetically encoded calcium probe Cameleon YC3.6 (Loro et al., 2012). To induce mitochondrial Ca2+ uptake, wild-type and knockout seedling roots were treated with external ATP as in previous studies (Loro et al., 2012) or auxin, known to induce cytosolic calcium increase in transition and elongation root tip cells (Monshausen et al., 2011). The ATP or auxin treatment produced mitochondrial Ca2+ uptake with peculiar dynamics for each treatment (Supplemental Fig. S4, A and E) in both genotypes, with no significant differences in the maximum mitochondrial calcium concentration ([Ca2+]mit) reached and the kinetics of accumulation and release (Supplemental Fig. S4). Moreover, a comparison of resting [Ca2+]mit in both genotypes did not show substantial differences (data not shown). Overall, in these young plants, no significant differences could be observed in mitochondrial calcium uptake or release. However, in 4-week-old plants, a significant reduction in the maximum [Ca2+]mit was observed in the mutant leaves compared with wild-type plants challenged with wounding stress (Supplemental Fig. S5, A and B), a stimulus known to induce steep cytosolic calcium increases in the cells surrounding the damaged area (Beneloujaephajri et al., 2013; Steinhorst and Kudla, 2013). Nevertheless, it has to be pointed out that even if the maximum calcium peak was different, this was the only observable phenotype, since no differences were observed in the calcium recovery phase (Supplemental Fig. S5).
DISCUSSION
We present, to our knowledge, the first known findings that one of the members of the iGLRs, AtGLR3.5, is located in the bioenergetic organelle mitochondria in plants. AtGLR3.5 is transcribed into two different splicing variants, one encoding for a protein displaying a clear N-terminal mitochondrial targeting peptide (see Aramemnon database [http://aramemnon.uni-koeln.de]) and the other harboring a predicted chloroplast-targeting peptide (ChloroP). Indeed, localization experiments show a dual targeting of AtGLR3.5 when expressed in a homologous system (i.e. agroinfiltrated Arabidopsis leaves). Several proteins are targeted to both mitochondria and chloroplasts because they have an ambiguous signal that can be recognized by both import systems (Silva-Filho, 2003). Dual targeting by ambiguous signals has been proposed to occur by combining distinct targeting instructions in a single N-terminal peptide (Karniely and Pines, 2005). Instead, we show that dual targeting of AtGLR3.5 is guided by alternative splicing. Please note that, due to the low abundance and high hydrophobicity, localization of ion channels in subcellular membranes by proteomic analysis proved to be feasible in only very few cases. To our knowledge, this is the first example of dual targeting achieved by this mechanism in the case of a plant ion channel. Among membrane proteins, a mitochondrial and chloroplastic dual-targeted ATP/ADP transporter of the mitochondrial carrier family in maize (Zea mays) and Arabidopsis, AtBT1 (for BRITTLE1), was shown to be localized to the inner plastidial envelope and mitochondria (Kirchberger et al., 2008; Bahaji et al., 2011). AtBT1, localized to the mitochondria, was found to be important for development and growth (Bahaji et al., 2011), but the function of the protein localized to the inner plastidial envelope remains unclear. Recently, a unique dual-targeted mitochondrial carrier protein, 3′-PHOSPHOADENOSINE 5′-PHOSPHOSULFATE TRANSPORTER1, was identified in rice (Oryza sativa) and its role in chloroplast development was proven (Xu et al., 2013). In our case, while the lack of AtGLR3.5 leads to clear changes in mitochondrial physiology, chloroplasts of knockout plants are normally developed with intact and stacked thylakoids present, even in the plants (3 weeks old) where mitochondrial changes were already visible. Chloroplasts have been shown previously to harbor AtGLR3.4 (Teardo et al., 2011); therefore, this latter iGLR likely overtakes the function of the chloroplast isoform of AtGLR3.5. In mammalian systems, several ion channels are located in multiple membranes within the cell (Köttgen et al., 2005; Shoshan-Barmatz and Israelson, 2005; Szabò and Zoratti, 2014). Furthermore, it is important to note that an N-methyl-d-aspartate Glu receptor has been found in the mitochondrial inner membrane in a hypothalamic neuronal cell line (Korde and Maragos, 2012).
Our results, using knockout and silenced Arabidopsis, provide evidence for the role of AtGLR3.5 in plant physiology. In particular, mitochondria lacking this putative channel show a strikingly compromised ultrastructure with altered cristae morphology and an overall increase in volume. Ion channels in the inner mitochondrial membrane are known to impact mitochondrial volume (Szabò and Zoratti, 2014). Therefore, the observed mitochondrial shape in the knockout plants suggests a localization of AtGLR3.5 in the inner membrane. Experiments using GLR3.5-SypHer are also indicative of such localization. These experiments also suggest that the Glu-recognizing part of the channel is oriented versus the cytosol. In both organelles, solute pores in the outer membranes are likely to mediate the flux of Glu, an important metabolite, thereby allowing the binding of Glu to inner mitochondrial membrane GLR3.5 and regulation of its activity. Our data suggest that the lack of this channel causes impairment in ion homeostasis that leads to a stress-like response and mitochondrial swelling. A similar mitochondrial ultrastructure has been observed in different plant species subjected to anaerobic treatment (Vartapetian et al., 2003). In this case, the authors interpreted their findings as the consequence of decreased metabolism (i.e. oxidative phosphorylation). Swelling was observed also upon treatment of Arabidopsis leaves with agents known to cause oxidative stress (hydrogen peroxide, paraquat, and menadione; Yoshinaga et al., 2005). Similarly, swollen mitochondria with altered cristae structure have been observed in chilled cucumber (Cucumis sativus) roots (Lee et al., 2002). Chill-sensitive plants undergo excessive oxidative stress, which, according to the interpretation of the authors, causes the ultrastructural changes observed in mitochondria. In this context, the observed accelerated senescence in 6-week-old plants might be a consequence of impaired mitochondrial integrity. The observation that GLR3.5 transcript level increases in older wild-type plants might indicate that AtGLR3.5 is required to maintain mitochondrial integrity and ATP production until late phases of senescence. It will be possible to clarify the exact link between the lack of AtGLR3.5, the mitochondrial changes, and the anticipated senescence only once the nature of the ions flowing through this protein and the regulation of channel activity are understood. Unfortunately, as mentioned, our attempts to perform patch-clamp experiments on isolated Arabidopsis mitochondria using a protocol that was previously successfully applied for wheat (Triticum aestivum) germ mitochondria (De Marchi et al., 2010) failed. In addition, attempts to record channel activity using the cell-free expressed protein incorporated into an artificial membrane performed as described previously (De Stefani et al., 2011) suggest that an additional, unknown component is necessary to reconstitute the functional AtGLR3.5 channel.
Because the previously studied members of the plant iGLRs of family 3 are permeable to Ca2+, we measured calcium dynamics with the genetically encoded calcium probe Cameleon targeted to mitochondria in wild-type and knockout plants. A significant difference in the maximum mitochondrial calcium accumulation could be observed only in 4-week-old plants challenged with wounding (used here just as a stimulus known to induce a steep cytosolic calcium increase in intact unstressed leaves). Nevertheless, the maximum peak was indeed the only difference, since the calcium recovery phase was not affected by the absence of GLR3.5. This might be a priori directly related to the lack of AtGLR3.5 but also might be due to an indirect effect (e.g. to a decreased driving force for the flux of calcium toward the matrix). Please note that other calcium-uptake pathways are likely to exist in mitochondria (Stael et al., 2012), including several isoforms of the mitochondrial calcium uniporter. Unfortunately, no experimental evidence is available about these alternative calcium-uptake pathways. However, it cannot be excluded that, in the early stages of plant development, the lack of AtGLR3.5 is counterbalanced by mitochondrial calcium uniporter function. Alternatively, the function of AtGLR3.5 for mitochondrial physiology might be decisive in a more advanced state of development and/or under stress conditions like those linked to aging or wounding. Swollen mitochondria, however, cannot be isolated, due to high fragility, in order to perform classical bioenergetics to assess mitochondrial physiology parameters. The observation that the expression of AtGLR3.5 increases with aging would be in accordance with the above hypothesis. In this respect, it is interesting to mention the case of stress-induced yeast (Saccharomyces cerevisiae) ortholog of the mammalian kidney disease gene Mpv17 (Sym1), a channel-forming protein (Reinhold et al., 2012) that is the yeast homolog of MPV17, a mammalian mitochondrial inner membrane protein, whose mutations are associated with and inherited autosomal mitochondrial disease characterized by neuronal disorders, growth retardation, liver failure, and hypoglycemia (e.g. Szabò and Zoratti, 2014). Sym1 knockout yeast cells show swollen mitochondria and lack of cristae especially under stress conditions, and stress in turn induces the expression of Sym1 in wild-type cells (Dallabona et al., 2010). While further work is warranted to identify the primary role of AtGLR3.5, our results indicate that, similar to Sym1, this iGLR is involved in the structural and functional stability of the inner mitochondrial membrane, thus controlling crucial mechanisms related to this compartment, including the morphology of mitochondria and the maintenance of its integrity. Furthermore, it is clear that the lack of this protein significantly impacts the physiology of the whole plant on a long-term scale, causing mitochondrial impairment and, possibly as a consequence, an anticipated senescence.
In summary, to our knowledge, this work provides evidence for the first molecular identification of a mitochondrial ion channel and provides insight into its physiological role.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) plants used for the experiments were grown in a growth room with controlled climatic conditions (16-h-light/8-h-dark photoperiod; relative humidity, 70%; night temperature, 18°C; and day temperature, 20°C). Seeds of Arabidopsis ecotype Col-0 were sterilized, incubated for 3 d at 4°C in the dark, and allowed to germinate in Murashige and Skoog (MS) medium (Duchefa) and solid medium (0.8% plant agar; Duchefa) supplemented with 30 g L−1 Suc in a growth chamber (20°C, 16-h-light/8-h-dark photoperiod, and 70% relative humidity). Seedlings were picked and grown in soil in a growth chamber (20°C, 16-h-light/8-h-dark photoperiod, and 70% relative humidity). Rosette leaves from 2-week-old plants were harvested and immediately powdered in liquid nitrogen for total RNA extraction. For genotyping, Arabidopsis wild-type (Col-0) and mutant plants were grown in a controlled growth chamber (20°C/18°C, 16-h-light/8-h-dark photoperiod, 100 μmol photons m−2 s−1 light, and 70% relative humidity). Mutant plants were transfer DNA insertion lines from the SALK collection: atglr3.5-1 (N656359, SALK_035264C) and atglr3.5-2 (N661846, SALK_023880C). After genomic DNA extraction using a standard protocol, mutants were genotyped by PCR using the forward and reverse primers listed in Supplemental Table S1.
Posttranscriptional Silencing of the GLR3.5 Gene
Arabidopsis wild-type plants were transformed with Agrobacterium tumefaciens carrying the pBIN Rolc plasmid containing a 204-bp-long sequence of the GLR3.5 mRNA linked to an antisense sequence corresponding to the same mRNA fragment (Molesini et al., 2009). Seeds collected from these plants were tested for posttranscriptional silencing of the GLR3.5 gene by extraction of mRNA from 4-week-old leaves.
Purification of Mitochondria
Leaves from 4-week-old Arabidopsis plants were homogenized in buffer A (330 mm Suc, 50 mm MOPS/KOH, pH 7.5, 5 mm EDTA, 5 mm dithiothreitol, 0.6% [w/v] polyvinylpolypyrrolidone, and 5 mm ascorbate) and filtered through a 250-μm nylon mesh. After centrifugation (10 s at 3,000g at 4°C), the supernatant was centrifuged for 15 min at 10,000g to collect the mitochondria (De Marchi et al., 2010).
Cloning, Subcellular Localization, and Topology
Total RNA was extracted from 100 mg of powdered leaves using the Trizol reagent (Gibco). After treatment with RNase-free DNase I (Ambion), first-strand cDNA was synthesized starting from 5 μg of total RNA using PowerScript Reverse Transcriptase (Clontech). The coding sequences of the two isoforms were amplified by PCR and cloned into the following vectors: G35v1 and G35v2 were amplified with primers G35v1_EcoRV_for/G35_SmaI_rev and G35v2_EcoRV_for/G35_SmaI_rev, respectively; PCR products were digested with EcoRV/SmaI and cloned in pGREAT::EGFP/DsRed2 vector (Carraretto et al., 2013; SmaI digested) for confocal microscopy; G35v1 was amplified with primers G35v1_NotI_for and G35v1_NotI_rev, digested with NotI, and cloned in pIVEX1.4 WG (Roche) vector for in vitro expression. Primers are listed in Supplemental Table S1. Enzymes were from New England Biolabs.
The subcellular localization of the AtGLR3.5 isoforms was examined through the agroinfiltration of 4-week-old Arabidopsis Col-0 wild-type leaves. Following infiltration, the plants were transferred to a growth room with controlled climatic conditions and examined to detect transformation after 4 d using a confocal microscope (Leica TCS SP5 II; Leica Microsystems). Images were collected with Leica Application Suite software. Chlorophyll and enhanced GFP were excited at 488 nm, while DsRed2 was excited at 543 nm. For fluorescence emission, we used wavelengths of 515 to 525, 570 to 620, and 680 to 720 nm for enhanced GFP, DsRed2, and chlorophyll, respectively.
SypHer was subcloned from the vector described by Poburko et al. (2011) into pGREAT-G35v1::DsRed2 to substitute the DsRed2 protein and obtain the pGREAT-G35v1::SypHer construct. Ratiometric sequential images of the 535-nm emission fluorescence in GLR3.5-SypHer-transformed (with polyethylene glycol) Arabidopsis protoplasts were acquired every 10 s during 30 min with a 63× objective of a Leica SP5 confocal microscope with alternative sequential excitation for 100 ms at 405 and 488 nm. As a control of acidification, cells were treated with 1 μm nigericin. The Multimeasure plug-in of ImageJ software was used to estimate mean fluorescence ratios of selected regions of interest (ROIs) matching mitochondria in at least four experiments following background subtraction; results are expressed as mtSypHer (405/488 nm) ratio.
Sequence Analysis
Bioinformatics analysis was performed using ChloroP and TargetP tools (http://www.cbs.dtu.dk/services), PCLRv_0.9 (http://andrewschein.com/pclr/), as well as BLASTP (http://www.ncbi.nih.gov).
Real-Time PCR
TaqMan assay amplification was performed in 1× TaqMan Universal PCR Master Mix (Applied Biosystems) using 1× final Custom TaqMan Assays. The thermal cycling conditions were 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. For every experiment, 100 ng of cDNA was used. Data were collected using the 7500 Real Time PCR system (Applied Biosystems). Each sample was tested in four replicates. Primers and TaqMan probes (Supplemental Table S2) were designed using Primer Express 1.5 software (Applied Biosystems) and purchased from Life Technologies.
TEM on Leaf Samples
A TEM investigation on leaf samples was conducted as described by Carraretto et al. (2013).
Seedling Preparation for Ca2+ Imaging
Arabidopsis seeds were surface sterilized by vapor-phase sterilization and plated on one-half-strength MS medium (MS salts, M0222 elements including vitamins [Duchefa; http://www.duchefa-biochemie.nl/]) supplemented with 0.1% (w/v) Suc and 2.34 mm MES, pH 6, and solidified with 0.8% (w/v) plant agar (Duchefa). After stratification at 4°C in the dark for 3 d, seeds were transferred to the growth chamber with 16-h-light/8-h-dark cycles (70 μmol m−2 s−1) at 20°C. The plates were kept vertically. Seedlings used for the analyses were 7 to 8 d old, which corresponds to an average root length of 3 cm. For root cell imaging, the seedlings were prepared according to Behera and Kudla (2013) in dedicated chambers and overlaid with wet cotton to continuously perfuse the root with the imaging solution (5 mm KCl, 10 mm MES, 10 mm Ca2+, pH 5.8, adjusted with Tris). The shoot was not submerged in the solution. ATP (final concentration of 0.1 mm) was added as disodium salt to the chamber by perfusion with the same solution. Similarly, the synthetic auxin naphthaleneacetic acid was added to the perfusion chamber with a final concentration of 0.01 mm.
Time-Lapse Ca2+ Imaging Analyses
Whole leaves of 4-week-old plants and seedling roots of both genotypes (wild-type-4mt-YC3.6 and AtGLR3.5-4mtYC3.6) expressing the mitochondria-localized Cameleon (Loro et al., 2012) were imaged in vivo by an inverted fluorescence microscope (Nikon Ti-E; Nikon; http://www.nikon.com/) with a CFI Planfluor 4×, numerical aperture 0.13 dry objective for entire leaves and CFI PLAN APO 20×, violet-corrected dry objective for roots. Excitation light was produced by a fluorescent lamp (Prior Lumen 200 PRO; Prior Scientific; http://www.prior.com) at 440 nm (436/20 nm) set to 50% (for leaves) or 20% (for roots). Images were collected with a Hamamatsu Dual CCD Camera (ORCA-D2; Hamamatsu Photonics; http://www.hamamatsu.com/). For Cameleon analysis, the fluorescence resonance energy transfer (FRET) cyan fluorescent protein (CFP)/yellow fluorescent protein optical block A11400-03 (emission 1, 483/32 nm for CFP; emission 2, 542/27 nm for FRET) with a dichroic 510-nm mirror (Hamamatsu Photonics) was used for the simultaneous CFP and cpVenus acquisitions. Exposure times were 500 ms for leaves and 100 ms for roots with a four-by-four CCD binning. Images were acquired every 2 and 5 s for leaves and roots, respectively. Filters and dichroic mirrors were purchased from Chroma Technology (http://www.chroma.com/). The NIS-Element (Nikon; http://www.nis-elements.com/) was used as a platform to control microscope, illuminator, camera, and postacquisition analyses. We used forceps for wounding as described in previous studies (Beneloujaephajri et al., 2013). Regarding time-course experiments, fluorescence intensity was determined over ROIs, which correspond to the root tip zone or the region surrounding the leaf wounded area. cpVenus and CFP emissions of the analyzed ROIs were used for the ratio (R) calculation (cpVenus/CFP) and normalized to the initial ratio (R0) and plotted versus time (ΔR/R0). Background subtraction was performed in each channel before FRET ratio calculation by selecting a ROI outside the sample, in the case of root analyses. In the case of analyses performed on leaves because imaged areas do not contain any background, the latter was not subtracted.
Semiquantitative RT-PCR
SAG12 (At5g45890) and AtGLR3.5 (At2g32390) gene expression was determined by semiquantitative RT-PCR using ACTIN2 (At3g18780) as an internal control. The PCR conditions were as follows: 10 s at 95°C, 30 s at 61°C, and 1 min at 72°C for 21 and 26 cycles for ACTIN2, SAG12, and AtGLR3.5 (both isoforms), respectively. Gel images were analyzed using the Geldoc system (Bio-Rad).
TMRM Staining
Sterilized seeds were sown on one-half-strength MS salts, 3% (w/v) Suc, 10 mm MES-KOH (pH 5.6), and 0.8% (w/v) plant agar (Micropoli) on sealed square plates. After 2 d at 4°C in the dark, seeds were incubated vertically in a growth chamber at 20°C and a 16-h-light/8-h-dark photoperiod for 8 d. Ten-day-old seedlings were incubated for 15 min in the imaging solution (10 mm MES, 5 mm KCl, and 10 mm CaCl2, pH 5.8) with 500 nm TMRM. After two washing steps in the imaging solution, the roots were viewed using a Leica TCS SP5 II confocal microscope (excitation, 543 nm; emission, 580–620 nm).
Chlorophyll Quantification and SDS-PAGE
Every measurement was performed on single leaves stored for 2 d in dimethylformamide at 4°C. Absorbance was measured at 647 and 664 nm, and chlorophyll was calculated as described by Carraretto et al. (2013). SDS-PAGE was performed in the presence of 6 m urea (Carraretto et al., 2013), while western blotting was carried out as described by Teardo et al. (2011). In vitro expression of GLR3.5 was performed as described by Carraretto et al. (2013). In Figure 2B, a 1-μL sample containing a maximum of 100 ng of the expressed protein was loaded; therefore, approximately 100 nmol (or less) of GLR3.5 protein was used.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The mitochondria isolated from Arabidopsis are enriched in cytochrome c.
Supplemental Figure S2. Transcript analysis of four putative silenced plants.
Supplemental Figure S3. Ultrastructure of organelles in wild-type, atglr3.5-1, and silenced plants.
Supplemental Figure S4. [Ca2+]mit dynamics in root tip cells of wild-type and atglr3.5-1 Arabidopsis seedlings expressing the 4mt-YC3.6 (mitochondrial Cameleon) probe.
Supplemental Figure S5. Mitochondrial calcium dynamics in leaf cells of wild-type and atglr3.5-1 plants.
Supplementary Table S1. Primers used for RT-PCR.
Supplementary Table S2. Primers and TaqMan probes used for real-time PCR.
Supplemental Movie S1. Arabidopsis leaves agroinfiltrated with A. tumefaciens strain harboring the pGREAT- 2x35S-G35v1::DsRed2 construct.
Supplemental Movie S2. Arabidopsis leaves agroinfiltrated with A. tumefaciens strain harboring the mito-mCherry construct that targets the mCherry reporter to the mitochondria.
Supplemental Movie S3. Ten-day-old Arabidopsis wild-type roots stained with TMRM.
Supplemental Movie S4. Ten-day-old Arabidopsis mutant (atglr3.5-1) roots stained with TMRM.
Supplementary Material
Acknowledgments
We thank Giorgio Mario Giacometti, Luca Scorrano, and Mario Zoratti for useful discussion. The SypHer probe and the silencing construct were kindly provided by Luca Scorrano and Angelo Spena, respectively.
Glossary
- iGluR
ionotropic glutamate receptor (animal)
- iGLR
ionotropic glutamate receptor (plant)
- NSCC
nonselective cation channel
- cDNA
complementary DNA
- RT
reverse transcription
- Col-0
Columbia-0
- TMRM
tetramethylrhodamine methyl ester
- TEM
transmission electron microscopy
- ROI
region of interest
- CFP
cyan fluorescent protein
Footnotes
This work was supported by Progetti di Ricerca di Interesse Nazionale (grant no. 2010CSJX4F_005 and Progetto di Ateneo 2009 to I.S.), by the Ministero dell’Istruzione, dell’Università e della Ricerca (grant no. RBFR10S1LJ_001 to A.C.), and by the University of Padova (postdoctoral fellowship to E.T.).
References
- Bahaji A, Muñoz FJ, Ovecka M, Baroja-Fernández E, Montero M, Li J, Hidalgo M, Almagro G, Sesma MT, Ezquer I, et al. (2011) Specific delivery of AtBT1 to mitochondria complements the aberrant growth and sterility phenotype of homozygous Atbt1 Arabidopsis mutants. Plant J 68: 1115–1121 [DOI] [PubMed] [Google Scholar]
- Behera S, Kudla J (July 1, 2013) High-resolution imaging of cytoplasmic Ca2+ dynamics in Arabidopsis roots. Cold Spring Harb Protoc 2013: http://dx.doi.org/10.1101/pdb.prot073023 [DOI] [PubMed] [Google Scholar]
- Beneloujaephajri E, Costa A, L’Haridon F, Métraux JP, Binda M (2013) Production of reactive oxygen species and wound-induced resistance in Arabidopsis thaliana against Botrytis cinerea are preceded and depend on a burst of calcium. BMC Plant Biol 13: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraretto L, Formentin E, Teardo E, Checchetto V, Tomizioli M, Morosinotto T, Giacometti GM, Finazzi G, Szabó I (2013) A thylakoid-located two-pore K+ channel controls photosynthetic light utilization in plants. Science 342: 114–118 [DOI] [PubMed] [Google Scholar]
- Chiu J, DeSalle R, Lam HM, Meisel L, Coruzzi G (1999) Molecular evolution of glutamate receptors: a primitive signaling mechanism that existed before plants and animals diverged. Mol Biol Evol 16: 826–838 [DOI] [PubMed] [Google Scholar]
- Chiu JC, Brenner ED, DeSalle R, Nitabach MN, Holmes TC, Coruzzi GM (2002) Phylogenetic and expression analysis of the glutamate-receptor-like gene family in Arabidopsis thaliana. Mol Biol Evol 19: 1066–1082 [DOI] [PubMed] [Google Scholar]
- Dallabona C, Marsano RM, Arzuffi P, Ghezzi D, Mancini P, Zeviani M, Ferrero I, Donnini C (2010) Sym1, the yeast ortholog of the MPV17 human disease protein, is a stress-induced bioenergetic and morphogenetic mitochondrial modulator. Hum Mol Genet 19: 1098–1107 [DOI] [PubMed] [Google Scholar]
- Davenport R. (2002) Glutamate receptors in plants. Ann Bot (Lond) 90: 549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marchi U, Checchetto V, Zanetti M, Teardo E, Soccio M, Formentin E, Giacometti GM, Pastore D, Zoratti M, Szabò I (2010) ATP-sensitive cation-channel in wheat (Triticum durum Desf.): identification and characterization of a plant mitochondrial channel by patch-clamp. Cell Physiol Biochem 26: 975–982 [DOI] [PubMed] [Google Scholar]
- Demidchik V, Essah PA, Tester M (2004) Glutamate activates cation currents in the plasma membrane of Arabidopsis root cells. Planta 219: 167–175 [DOI] [PubMed] [Google Scholar]
- Dennison KL, Spalding EP (2000) Glutamate-gated calcium fluxes in Arabidopsis. Plant Physiol 124: 1511–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Stefani D, Raffaello A, Teardo E, Szabò I, Rizzuto R (2011) A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476: 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingledine R, Borges K, Bowie D, Traynelis SF (1999) The glutamate receptor ion channels. Pharmacol Rev 51: 7–61 [PubMed] [Google Scholar]
- Dubos C, Huggins D, Grant GH, Knight MR, Campbell MM (2003) A role for glycine in the gating of plant NMDA-like receptors. Plant J 35: 800–810 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Gan S. (2007) Senescence Processes in Plants. Blackwell Publishing Ltd, Oxford [Google Scholar]
- Kang J, Mehta S, Turano FJ (2004) The putative glutamate receptor 1.1 (AtGLR1.1) in Arabidopsis thaliana regulates abscisic acid biosynthesis and signaling to control development and water loss. Plant Cell Physiol 45: 1380–1389 [DOI] [PubMed] [Google Scholar]
- Kang J, Turano FJ (2003) The putative glutamate receptor 1.1 (AtGLR1.1) functions as a regulator of carbon and nitrogen metabolism in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 6872–6877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Kim HB, Lee H, Choi JY, Heu S, Oh CJ, Kwon SI, An CS (2006) Overexpression in Arabidopsis of a plasma membrane-targeting glutamate receptor from small radish increases glutamate-mediated Ca2+ influx and delays fungal infection. Mol Cells 21: 418–427 [PubMed] [Google Scholar]
- Karniely S, Pines O (2005) Single translation-dual destination: mechanisms of dual protein targeting in eukaryotes. EMBO Rep 6: 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SA, Kwak JM, Jae SK, Wang MH, Nam HG (2001) Overexpression of the AtGluR2 gene encoding an Arabidopsis homolog of mammalian glutamate receptors impairs calcium utilization and sensitivity to ionic stress in transgenic plants. Plant Cell Physiol 42: 74–84 [DOI] [PubMed] [Google Scholar]
- Kirchberger S, Tjaden J, Neuhaus HE (2008) Characterization of the Arabidopsis Brittle1 transport protein and impact of reduced activity on plant metabolism. Plant J 56: 51–63 [DOI] [PubMed] [Google Scholar]
- Korde AS, Maragos WF (2012) Identification of an N-methyl-D-aspartate receptor in isolated nervous system mitochondria. J Biol Chem 287: 35192–35200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köttgen M, Benzing T, Simmen T, Tauber R, Buchholz B, Feliciangeli S, Huber TB, Schermer B, Kramer-Zucker A, Höpker K, et al. (2005) Trafficking of TRPP2 by PACS proteins represents a novel mechanism of ion channel regulation. EMBO J 24: 705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol E, Dziubinska H, Trebacz K, Koselski M, Stolarz M (2007) The influence of glutamic and aminoacetic acids on the excitability of the liverwort Conocephalum conicum. J Plant Physiol 164: 773–784 [DOI] [PubMed] [Google Scholar]
- Kwaaitaal M, Huisman R, Maintz J, Reinstädler A, Panstruga R (2011) Ionotropic glutamate receptor (iGluR)-like channels mediate MAMP-induced calcium influx in Arabidopsis thaliana. Biochem J 440: 355–365 [DOI] [PubMed] [Google Scholar]
- Lam HM, Chiu J, Hsieh MH, Meisel L, Oliveira IC, Shin M, Coruzzi G (1998) Glutamate-receptor genes in plants. Nature 396: 125–126 [DOI] [PubMed] [Google Scholar]
- Lee SH, Singh AP, Chung GC, Kim YS, Kong IB (2002) Chilling root temperature causes rapid ultrastructural changes in cortical cells of cucumber (Cucumis sativus L.) root tips. J Exp Bot 53: 2225–2237 [DOI] [PubMed] [Google Scholar]
- Li J, Zhu S, Song X, Shen Y, Chen H, Yu J, Yi K, Liu Y, Karplus VJ, Wu P, et al. (2006) A rice glutamate receptor-like gene is critical for the division and survival of individual cells in the root apical meristem. Plant Cell 18: 340–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loro G, Drago I, Pozzan T, Schiavo FL, Zottini M, Costa A (2012) Targeting of Cameleons to various subcellular compartments reveals a strict cytoplasmic/mitochondrial Ca2+ handling relationship in plant cells. Plant J 71: 1–13 [DOI] [PubMed] [Google Scholar]
- McAinsh MR, Pittman JK (2009) Shaping the calcium signature. New Phytol 181: 275–294 [DOI] [PubMed] [Google Scholar]
- Meyerhoff O, Müller K, Roelfsema MRG, Latz A, Lacombe B, Hedrich R, Dietrich P, Becker D (2005) AtGLR3.4, a glutamate receptor channel-like gene is sensitive to touch and cold. Planta 222: 418–427 [DOI] [PubMed] [Google Scholar]
- Michard E, Lima PT, Borges F, Silva AC, Portes MT, Carvalho JE, Gilliham M, Liu LH, Obermeyer G, Feijó JA (2011) Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil D-serine. Science 332: 434–437 [DOI] [PubMed] [Google Scholar]
- Molesini B, Pandolfini T, Rotino GL, Dani V, Spena A (2009) Aucsia gene silencing causes parthenocarpic fruit development in tomato. Plant Physiol 149: 534–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen GB, Miller ND, Murphy AS, Gilroy S (2011) Dynamics of auxin-dependent Ca2+ and pH signaling in root growth revealed by integrating high-resolution imaging with automated computer vision-based analysis. Plant J 65: 309–318 [DOI] [PubMed] [Google Scholar]
- Mousavi SA, Chauvin A, Pascaud F, Kellenberger S, Farmer EE (2013) GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature 500: 422–426 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Poburko D, Santo-Domingo J, Demaurex N (2011) Dynamic regulation of the mitochondrial proton gradient during cytosolic calcium elevations. J Biol Chem 286: 11672–11684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Stephens NR, Spalding EP (2006) Calcium entry mediated by GLR3.3, an Arabidopsis glutamate receptor with a broad agonist profile. Plant Physiol 142: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold R, Krüger V, Meinecke M, Schulz C, Schmidt B, Grunau SD, Guiard B, Wiedemann N, van der Laan M, Wagner R, et al. (2012) The channel-forming Sym1 protein is transported by the TIM23 complex in a presequence-independent manner. Mol Cell Biol 32: 5009–5021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R, De Stefani D, Raffaello A, Mammucari C (2012) Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol 13: 566–578 [DOI] [PubMed] [Google Scholar]
- Scott I, Logan DC (2008) Mitochondrial morphology transition is an early indicator of subsequent cell death in Arabidopsis. New Phytol 177: 90–101 [DOI] [PubMed] [Google Scholar]
- Shoshan-Barmatz V, Israelson A (2005) The voltage-dependent anion channel in endoplasmic/sarcoplasmic reticulum: characterization, modulation and possible function. J Membr Biol 204: 57–66 [DOI] [PubMed] [Google Scholar]
- Silva-Filho MC. (2003) One ticket for multiple destinations: dual targeting of proteins to distinct subcellular locations. Curr Opin Plant Biol 6: 589–595 [DOI] [PubMed] [Google Scholar]
- Sivaguru M, Pike S, Gassmann W, Baskin TI (2003) Aluminum rapidly depolymerizes cortical microtubules and depolarizes the plasma membrane: evidence that these responses are mediated by a glutamate receptor. Plant Cell Physiol 44: 667–675 [DOI] [PubMed] [Google Scholar]
- Stael S, Wurzinger B, Mair A, Mehlmer N, Vothknecht UC, Teige M (2012) Plant organellar calcium signalling: an emerging field. J Exp Bot 63: 1525–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhorst L, Kudla J (2013) Calcium and reactive oxygen species rule the waves of signaling. Plant Physiol 163: 471–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens NR, Qi Z, Spalding EP (2008) Glutamate receptor subtypes evidenced by differences in desensitization and dependence on the GLR3.3 and GLR3.4 genes. Plant Physiol 146: 529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabò I, Zoratti M (2014) Mitochondrial channels: ion fluxes and more. Physiol Rev 94: 519–608 [DOI] [PubMed] [Google Scholar]
- Tapken D, Anschütz U, Liu LH, Huelsken T, Seebohm G, Becker D, Hollmann M (2013) A plant homolog of animal glutamate receptors is an ion channel gated by multiple hydrophobic amino acids. Sci Signal 6: ra47. [DOI] [PubMed] [Google Scholar]
- Tapken D, Hollmann M (2008) Arabidopsis thaliana glutamate receptor ion channel function demonstrated by ion pore transplantation. J Mol Biol 383: 36–48 [DOI] [PubMed] [Google Scholar]
- Teardo E, Formentin E, Segalla A, Giacometti GM, Marin O, Zanetti M, Lo Schiavo F, Zoratti M, Szabò I (2011) Dual localization of plant glutamate receptor AtGLR3.4 to plastids and plasmamembrane. Biochim Biophys Acta 1807: 359–367 [DOI] [PubMed] [Google Scholar]
- Teardo E, Segalla A, Formentin E, Zanetti M, Marin O, Giacometti GM, Lo Schiavo F, Zoratti M, Szabò I (2010) Characterization of a plant glutamate receptor activity. Cell Physiol Biochem 26: 253–262 [DOI] [PubMed] [Google Scholar]
- Vartapetian BB, Andreeva IN, Generozova IP, Polyakova LI, Maslova IP, Dolgikh YI, Stepanova AY (2003) Functional electron microscopy in studies of plant response and adaptation to anaerobic stress. Ann Bot (Lond) 91: 155–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincill ED, Bieck AM, Spalding EP (2012) Ca2+ conduction by an amino acid-gated ion channel related to glutamate receptors. Plant Physiol 159: 40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincill ED, Clarin AE, Molenda JN, Spalding EP (2013) Interacting glutamate receptor-like proteins in phloem regulate lateral root initiation in Arabidopsis. Plant Cell 25: 1304–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Liu P, Liu LH, Remans T, Tester M, Forde BG (2006) Evidence that L-glutamate can act as an exogenous signal to modulate root growth and branching in Arabidopsis thaliana. Plant Cell Physiol 47: 1045–1057 [DOI] [PubMed] [Google Scholar]
- Xu J, Yang J, Wu Z, Liu H, Huang F, Wu Y, Carrie C, Narsai R, Murcha M, Whelan J, et al. (2013) Identification of a dual-targeted protein belonging to the mitochondrial carrier family that is required for early leaf development in rice. Plant Physiol 161: 2036–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga K, Arimura S, Niwa Y, Tsutsumi N, Uchimiya H, Kawai-Yamada M (2005) Mitochondrial behaviour in the early stages of ROS stress leading to cell death in Arabidopsis thaliana. Ann Bot (Lond) 96: 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.