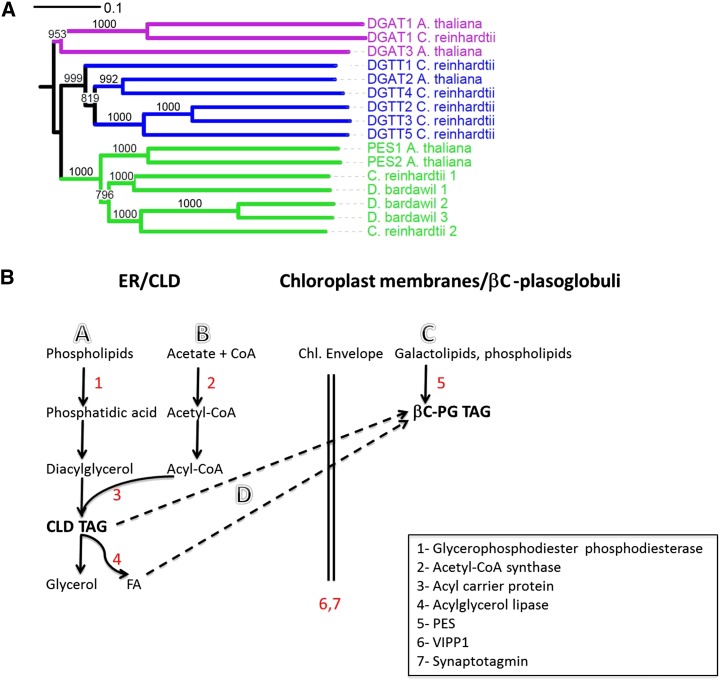
Figure 6.
Phylogenetic tree of PES and DGAT in βC-plastoglobuli and schemes of proposed TAG biosynthesis in CLD and βC-plastoglobuli. A, Phylogenic tree of PES and DGAT showing that the three isotigs in D. bardawil βC-plastoglobuli show higher homology to PES than to DGAT. D. bardawil 1 to 3 refer to isotigs isotig15851, CL1Contig10166.1, and isotig04015, respectively. Sequence names followed by organism and NCBI accession numbers are as follows: PES1_AT1G54570 (Arabidopsis; NP_564662.1), PES2_AT3G26840 (Arabidopsis; NP_566801.1), DGAT1 (Arabidopsis; NP_179535.1), DGAT2_AT3G51520 (Arabidopsis; NP_566952.1), DGAT1_Chlamy (C. reinhardtii; from Boyle et al. [2012]), DGTT1_Chlamy (C. reinhardtii; AFB73929.1), DGTT2_Chlamy (C. reinhardtii; XM_001694852), DGTT4_Chlamy (C. reinhardtii; XM_001693137), DGTT5_Chlamy (C. reinhardtii; XM_001701615), Chlamy_Cre08.g365950 (C. reinhardtii; XP_001696047.1), Chlamy_Cre12.g521650 (C. reinhardtii; XP_001696915.1). B, Proposed TAG biosynthesis and mobilization in CLD and βC-plastoglobules in D. bardawil. Paths are as follows: A, fatty acid (FA) recycling from phospholipids; B, de novo synthesis; C, fatty acid recycling from galactolipids; and D, CLD TAG recycling.